
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,087,614 – 6,087,714 |
| Length | 100 |
| Max. P | 0.512930 |

| Location | 6,087,614 – 6,087,714 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.16 |
| Shannon entropy | 0.57594 |
| G+C content | 0.37329 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -4.11 |
| Energy contribution | -4.21 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.512930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
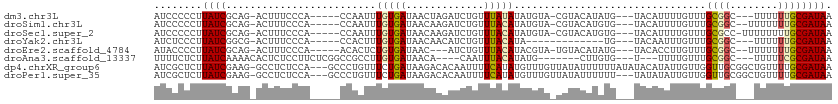
>dm3.chr3L 6087614 100 - 24543557 AUCCCCCUUAUCGCAG-ACUUUCCCA-----CCAAUUUGUGAUAACUAGAUCUGUUUAUAUAUGUA-CGUACAUAUG---UACAUUUUGUUUGCGGC---UUUUUUGCGAUAA ........((((((((-(......((-----(......)))......(((.((((..(((.(((((-(((....)))---)))))..)))..)))).---)))))))))))). ( -23.60, z-score = -4.00, R) >droSim1.chr3L 5590693 101 - 22553184 AUCCCCCUUAUCGCAG-ACUUUCCCA-----CCAAUUUGUGAUAACAAGAUCUGUUUACAUAUGUA-CGUACAUGUG---UACAUUUUGUUUGCGGC--UUUUUUUGCGAUAA ........((((((((-(......((-----(......))).....((((.((((..(((.(((((-((......))---)))))..)))..)))).--))))))))))))). ( -25.20, z-score = -3.89, R) >droSec1.super_2 6035962 102 - 7591821 AUCCCCCUUAUCGCAG-ACUUUCCCA-----CCAAUUUGUGAUAACAAGAUCUGUUUACAUAUGUA-CGUACAUGUG---UACAUUUUGUUUGCGCC-UUUUUUUUGCGAUAA ........((((((((-(......((-----(......)))..((((((((.....(((((((((.-...)))))))---)).))))))))......-.....))))))))). ( -23.60, z-score = -4.32, R) >droYak2.chr3L 6670269 88 - 24197627 AUCUCCCUUAUCGGCG-ACUUUCCCA-----CCACUUUGUGAUAACAACAUCUGUUUACAUA-------------UG---UACAAUUUGUUUGCGGC---UUUUUUGCGAUAA ........(((((.((-(........-----.....((((....))))...((((..(((.(-------------(.---....)).)))..)))).---....)))))))). ( -11.90, z-score = -0.19, R) >droEre2.scaffold_4784 8784835 98 - 25762168 AUACCCCUUAUCGCAG-ACUUUCCCA-----ACACUCUGUGAUAAC---AUCUGUUUACAUACGUA-UGUACAUAUG---UACACCUUGUUUGCGGC--UUUUUUUGCGAUAA ........((((((((-(....((((-----(.((...(((...((---((.(((.(((....)))-...))).)))---).)))...))))).)).--....))))))))). ( -20.90, z-score = -3.03, R) >droAna3.scaffold_13337 2439790 93 + 23293914 UUUUCUCUUAUCAAAACACUCUCCUUCUCGGCCGCCUUGUGAUAACA----CAAUUUACAUAUG-------CUUGUG---U---UUUUGUUUGCGGC---UUUUUCGCGAUAA .............................((((((...(..(.((((----(((..........-------.)))))---)---).)..)..)))))---)............ ( -17.30, z-score = -2.12, R) >dp4.chrXR_group6 3724876 109 + 13314419 AUCGCUCUUAUCGAAG-GCCUCUCCA---GCCCUGUUUCUGAUAAGACACAAUUUUCAUAUGUUUGUUAUAUUUUUUAUAUACAUAUUGUUGGUUGCGGCUGUUUUGCGAUAA ((((((((((((((((-((.......---)))....))).)))))))(.(((((..(((((((....((((.....)))).))))).))..))))).)........))))).. ( -24.80, z-score = -2.14, R) >droPer1.super_35 134854 106 - 973955 AUCGCUCUUAUCGAAG-GCCUCUCCA---GCCCUGUUUCUGAUAAGACACAAUUUUCAUAUGUUUGUUAUAUUUUUU---UAUAUAUUGUUGGUUGCGGCUGUUUUGCGAUAA (((((((.....)).(-(((.(.(((---((..(((.(((....))).)))................(((((.....---.)))))..)))))..).)))).....))))).. ( -21.30, z-score = -1.21, R) >consensus AUCCCCCUUAUCGCAG_ACUUUCCCA_____CCAAUUUGUGAUAACAAGAUCUGUUUACAUAUGUA_CGUACAUGUG___UACAUUUUGUUUGCGGC___UUUUUUGCGAUAA ........((((.........................(((((.............)))))................................((((........)))))))). ( -4.11 = -4.21 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:33 2011