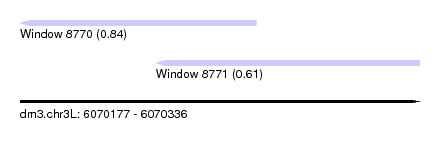
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,070,177 – 6,070,336 |
| Length | 159 |
| Max. P | 0.841683 |

| Location | 6,070,177 – 6,070,271 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Shannon entropy | 0.44486 |
| G+C content | 0.68412 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -26.83 |
| Energy contribution | -27.94 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
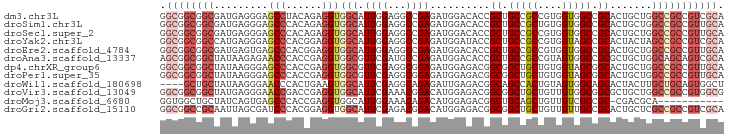
>dm3.chr3L 6070177 94 - 24543557 GGCGGCGGCGAUGAGGGAGCCUACAGAGGUGGCAUUGGAGGCCGAGAUGGACACCGCUGCCGCCGUGGUGGCCGCACUGCUGGCCGCCGUCGCA .(((((((((........(((..(((.(((((((.(((.(.((.....)).).))).)))))))((((...)))).)))..)))))))))))). ( -48.30, z-score = -1.37, R) >droSim1.chr3L 5572919 94 - 22553184 GGCGGCGGCGAUGAGGGAGCCCACAGAGGUGGCAUUGGAGGCCGAGAUGGACACCGCUGCCGCUGUGGUGGCCGCACUGCUGGCCGCCGUUGCA ((((((((.(....((....)).((....((((.......))))...))..).))))))))((..((((((((((...)).))))))))..)). ( -46.20, z-score = -0.98, R) >droSec1.super_2 6018695 94 - 7591821 GGCGGCGGCGAUGAGGGAGCCCACAGAGGUGGCAUUGGAGGCCGAGAUGGACACCGCUGCCGCCGUGGUGGCCGCACUGCUGGCCGCCGUUGCA ((((((((((.((.((....)).((....((((.......))))...))..)).))))))))))..(((((((((...)).)))))))...... ( -48.10, z-score = -1.34, R) >droYak2.chr3L 6652099 94 - 24197627 GGCGGCGGCCAUGAGGGAGCCCACGGAGGUGGCAUUGGAGGCCGAGAUGGAUACCGCUGCCGCCGUGGUAGCCGCACUACUAGCCGCCGUCGCA (((((((((.....((....)).....(((((((.(((...((.....))...))).))))))).((((((.....)))))))))))))))... ( -45.90, z-score = -1.26, R) >droEre2.scaffold_4784 8768123 94 - 25762168 GGCGGCGGCGAUGAGUGAGCCCACGGAGGUGGCAUUGGAGGCCGAGAUGGACACCGCUGCCGCCGUGGUGGCCGCACUGCUGGCCGCCGUUGCA (((((((((.(..((((.(.((((.(.(((((((.(((.(.((.....)).).))).))))))).).)))).).))))..).)))))))))... ( -50.60, z-score = -2.00, R) >droAna3.scaffold_13337 2422704 94 + 23293914 AGCGGCGGCUAUAAGAGAACCCACCGAGGUGGCGUUCGAUGCCGAGAUGGACACCGCUGCCGCCGUAGUGGCCGCGCUGCUGGCAGCAGUCGCA .((((((((.((....((((((((....)))).)))).)))))............(((((((..((((((....))))))))))))).))))). ( -44.90, z-score = -1.73, R) >dp4.chrXR_group6 3707326 94 + 13314419 GGCGGCGGCUAUAAGGGAGCCCACCGAGGUGGCGUUCGAGGCGGAGAUGGAGACGGCGGCUGCUGUGGUAGCGGCACUGCUGGCCGCCGUUGCA ((((((((((......((((((((....)))).))))..(((((...((....))((.(((((....))))).)).)))))))))))))))... ( -50.90, z-score = -2.90, R) >droPer1.super_35 117258 94 - 973955 GGCGGCGGCUAUAAGGGAGCCCACCGAGGUGGCGUUCGAGGCGGAGAUGGAGACGGCGGCUGCUGUGGUAGCGGCACUGCUGGCCGCCGUUGCA ((((((((((......((((((((....)))).))))..(((((...((....))((.(((((....))))).)).)))))))))))))))... ( -50.90, z-score = -2.90, R) >droWil1.scaffold_180698 3337325 90 - 11422946 ----GCUGCUAUAAGGGAACCCACUGAAGUGGCAUUCGAGGCAGAGAUUGAGACGGCAGCCACUGUAGUGGCAGCACUACUUGCUGCAGUGGCU ----((..((...........(((((.((((((..(((...(((...)))...)))..)))))).)))))((((((.....))))))))..)). ( -34.00, z-score = -1.29, R) >droVir3.scaffold_13049 574401 94 - 25233164 GGCGGCGGCUAUGAGGGAACCGACCGAGGUGGCAUUCGAAACGGACAUGGAGACGGCGGCUGCUGUUGUGGCGGCGCUGCUGGCCGCCGUGGCG ((((((.(((.........(((..((((......))))...)))....(....)))).))))))((..(((((((.(....))))))))..)). ( -42.60, z-score = -1.11, R) >droMoj3.scaffold_6680 24120913 82 + 24764193 GGUGGCUGCUAUCAGUGAGCCCACCGAGGUGGCAUUGGAAACAGACAUGGAGACGGCUGCAGCUGUUGUCGCCGC-CGACGCA----------- (((((((((.....)).)).)))))..((((((..((....))((((.....(((((....))))))))))))))-)......----------- ( -34.50, z-score = -1.45, R) >droGri2.scaffold_15110 1826173 94 - 24565398 GGCGGCCGCAAUUAGCGAUCCCACGGAGGUGGCAUUCGAGACGGACAUGGAGACGGCGGCUGCUGUUGUUGCCGCACUGCUCGCCGCCGUCGCA .((((((((.....))).(((...)))((((((....(((.(((...((....))(((((.((....)).))))).))))))))))))))))). ( -41.90, z-score = -0.69, R) >consensus GGCGGCGGCUAUGAGGGAGCCCACCGAGGUGGCAUUCGAGGCCGAGAUGGAGACCGCUGCCGCUGUGGUGGCCGCACUGCUGGCCGCCGUUGCA .((((((((.........(((......)))(((.((((...))))..........((.(((((....))))).)).......))))))))))). (-26.83 = -27.94 + 1.11)
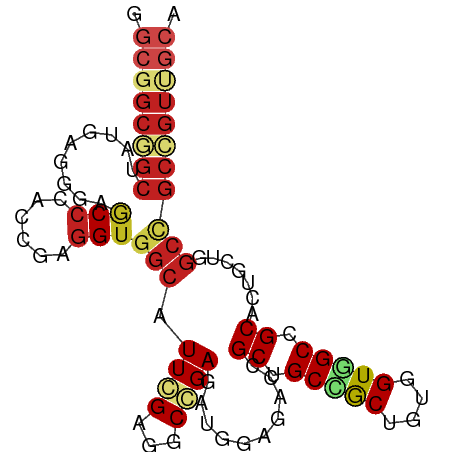

| Location | 6,070,231 – 6,070,336 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 63.10 |
| Shannon entropy | 0.69820 |
| G+C content | 0.62394 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -14.18 |
| Energy contribution | -13.70 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.607235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
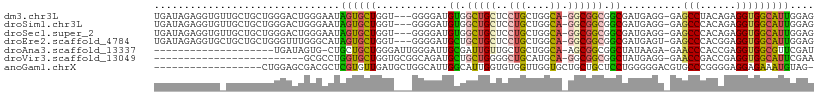
>dm3.chr3L 6070231 105 - 24543557 UGAUAGAGGUGUUGCUGCUGGGACUGGGAAUAGUGCUGGU---GGGGAUGUGGCUGCUCCUGCUGGCA-GGCGGCGGCGAUGAGG-GAGCCUACAGAGGUGGCAUUGGAG ........(..((.(..(..(.((((....)))).)..).---.).))..).((((((((((....))-)).))))))((((...-..((((....))))..)))).... ( -37.00, z-score = -1.02, R) >droSim1.chr3L 5572973 105 - 22553184 UGAUAGAGGUGUUGCUGCUGGGACUGGGAAUAGUGCUGGU---GGGGAUGUGGCUGCUCCUGCUGGCA-GGCGGCGGCGAUGAGG-GAGCCCACAGAGGUGGCAUUGGAG .......(((((..((((..(.((((....)))).)..))---.....(((((..((((((((((.(.-...).)))).....))-)))))))))..))..))))).... ( -38.80, z-score = -1.04, R) >droSec1.super_2 6018749 105 - 7591821 UGAUAGAGGUGUUGCUGCUGGGACUGGGAAUAGUGCUGGU---GGGGAUGUGGCUGCUCCUGCUGGCA-GGCGGCGGCGAUGAGG-GAGCCCACAGAGGUGGCAUUGGAG .......(((((..((((..(.((((....)))).)..))---.....(((((..((((((((((.(.-...).)))).....))-)))))))))..))..))))).... ( -38.80, z-score = -1.04, R) >droEre2.scaffold_4784 8768177 105 - 25762168 UGAUAGAGGUGCUGCUGCUGGGUUUGGGCAUAGUGCUGGU---GGGGAUGCUGCUGCUCCUGCUGGCA-GGCGGCGGCGAUGAGU-GAGCCCACGGAGGUGGCAUUGGAG .......(((((..((.((((((((.(((((..(.(....---).).)))))((((((((((....))-)).)))))).......-))))))).)..))..))))).... ( -40.50, z-score = 0.04, R) >droAna3.scaffold_13337 2422758 87 + 23293914 --------------------UGAUAGUG-CUGCUGCUGGGAUUGGGAUUGCGAUUGUUGCUGCUGGCA-AGCGGCGGCUAUAAGA-GAACCCACCGAGGUGGCGUUCGAU --------------------.......(-((((((((..(((((......)))))..(((.....)))-))))))))).......-((((((((....)))).))))... ( -32.10, z-score = -1.52, R) >droVir3.scaffold_13049 574455 83 - 25233164 -------------------------GCGCCUGGUGCUGGUGCGGCAGAUGCUGCUGGGGCUGCAUGCA-GGCGGCGGCUAUGAGG-GAACCGACCGAGGUGGCAUUCGAA -------------------------.((((((((((.(.(.((((((...)))))).).).)))).))-))))((.(((....((-.......))..))).))....... ( -34.30, z-score = 0.07, R) >anoGam1.chrX 19765741 91 - 22145176 ------------------CUGGAGCGACGCUCGUGUUGAUGCUGGCAUUGGCAUUGGUGUGGUUGGUGCUGCUGCUCCUGGGGGACGUGCCCGGGGAGGAGAAAUGUAG- ------------------....(((.(((((.((((..(((....)))..)))).))))).)))..(((..((.(((((.(((......))).))))).))....))).- ( -39.50, z-score = -3.15, R) >consensus UGAUAGAGGUG_UGCUGCUGGGACUGGGAAUAGUGCUGGU___GGGGAUGCGGCUGCUCCUGCUGGCA_GGCGGCGGCGAUGAGG_GAGCCCACAGAGGUGGCAUUGGAG ...............................((((((............((.(((((...((....))..))))).))..........(((......))))))))).... (-14.18 = -13.70 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:31 2011