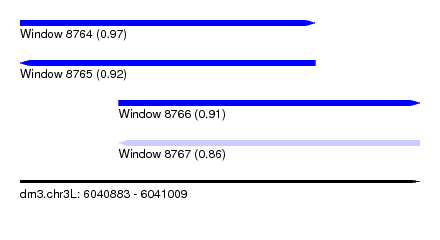
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,040,883 – 6,041,009 |
| Length | 126 |
| Max. P | 0.970232 |

| Location | 6,040,883 – 6,040,976 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.76 |
| Shannon entropy | 0.13261 |
| G+C content | 0.43728 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
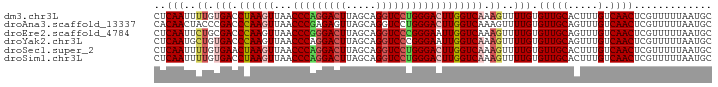
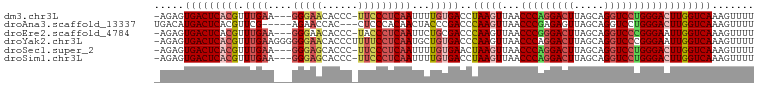
>dm3.chr3L 6040883 93 + 24543557 GCAUUAAAAACGAGUUGACAAAGUGCAACACAAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGGUCACAAAAUUGAG .............((((.(.....))))).(((.....(((((((((((((((((.......))))))))...)))).))))).....))).. ( -27.20, z-score = -3.07, R) >droAna3.scaffold_13337 16725997 93 + 23293914 GCAUUAAAAACGAGUUGACAAACUGCAACACAAAACUUUGACCAAGUCCCAGGACCUGCUAACUCUCGGGUUAACUUGGGUCGGGUAGUUGUG ............((((....))))(((((.(((....)))(((....(((((((((((........)))))...))))))...))).))))). ( -23.10, z-score = -0.49, R) >droEre2.scaffold_4784 8738061 93 + 25762168 GCAUUAAAAACGAGUUGACAAACUGCAACACAAAACUUUGACCAAUUCCCGGGACCUGCUAAGUCCCGGGUUAACUUGGGUCGCAGAAUUGAG .............((((.(.....))))).(((..((.(((((....((((((((.......))))))))........))))).))..))).. ( -27.50, z-score = -2.69, R) >droYak2.chr3L 6621913 93 + 24197627 GCAUUAAAAACGAGUUGACAAACUGCAACACAAAACUUUGACCAAUUCCCGGGACCUGCUAAGUCCUGGGUUAACUUGGGUCACAGCAUUGAG .............((((.(.....))))).(((..((.(((((....((((((((.......))))))))........))))).))..))).. ( -26.20, z-score = -2.29, R) >droSec1.super_2 5990649 93 + 7591821 GCAUUAAAAACGAGUUGACAAAGUGCAACACAAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGUUCACAAAAUUGAG ........((((((((((((((((..........)))))).......((((((((.......))))))))))))))).)))............ ( -24.20, z-score = -2.40, R) >droSim1.chr3L 5543072 93 + 22553184 GCAUUAAAAACGAGUUGACAAAGUGCAACACAAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGGUCACAAAAUUGAG .............((((.(.....))))).(((.....(((((((((((((((((.......))))))))...)))).))))).....))).. ( -27.20, z-score = -3.07, R) >consensus GCAUUAAAAACGAGUUGACAAACUGCAACACAAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGGUCACAAAAUUGAG .............((((.(.....))))).(((.....(((((((((((((((((.......))))))))...)))).))))).....))).. (-23.22 = -23.42 + 0.20)

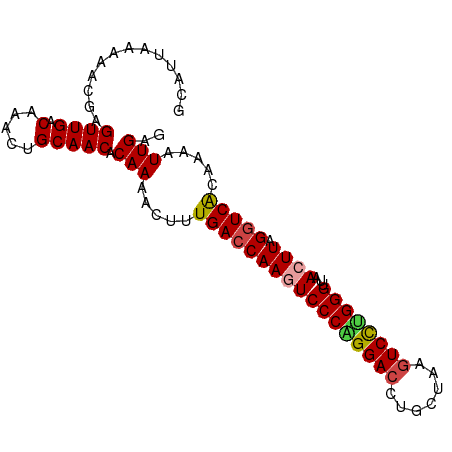
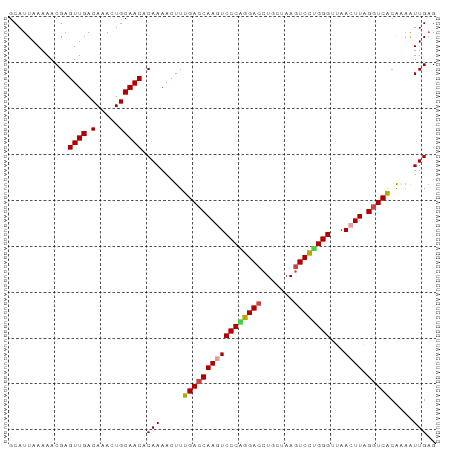
| Location | 6,040,883 – 6,040,976 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.76 |
| Shannon entropy | 0.13261 |
| G+C content | 0.43728 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -23.78 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6040883 93 - 24543557 CUCAAUUUUGUGACCUAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUUGUGUUGCACUUUGUCAACUCGUUUUUAAUGC ..(((..((.(((((.((((...(((((((((.....)))))))))))))))))).))..))).(((((.....).))))............. ( -29.80, z-score = -3.03, R) >droAna3.scaffold_13337 16725997 93 - 23293914 CACAACUACCCGACCCAAGUUAACCCGAGAGUUAGCAGGUCCUGGGACUUGGUCAAAGUUUUGUGUUGCAGUUUGUCAACUCGUUUUUAAUGC (((((..((..((((...((((((......))))))(((((....)))))))))...)).)))))..((((((....)))).))......... ( -19.50, z-score = 0.52, R) >droEre2.scaffold_4784 8738061 93 - 25762168 CUCAAUUCUGCGACCCAAGUUAACCCGGGACUUAGCAGGUCCCGGGAAUUGGUCAAAGUUUUGUGUUGCAGUUUGUCAACUCGUUUUUAAUGC ..(((..(((((((.(((.....(((((((((.....)))))))))((((......))))))).))))))).))).................. ( -30.40, z-score = -2.96, R) >droYak2.chr3L 6621913 93 - 24197627 CUCAAUGCUGUGACCCAAGUUAACCCAGGACUUAGCAGGUCCCGGGAAUUGGUCAAAGUUUUGUGUUGCAGUUUGUCAACUCGUUUUUAAUGC ..(((.(((.(((((..((((..(((.(((((.....))))).)))))))))))).))).))).(((((.....).))))............. ( -24.60, z-score = -1.04, R) >droSec1.super_2 5990649 93 - 7591821 CUCAAUUUUGUGAACUAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUUGUGUUGCACUUUGUCAACUCGUUUUUAAUGC ...(((((((...(((((((...(((((((((.....)))))))))))))))))))))))..(.(((((.....).)))).)........... ( -28.60, z-score = -2.75, R) >droSim1.chr3L 5543072 93 - 22553184 CUCAAUUUUGUGACCUAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUUGUGUUGCACUUUGUCAACUCGUUUUUAAUGC ..(((..((.(((((.((((...(((((((((.....)))))))))))))))))).))..))).(((((.....).))))............. ( -29.80, z-score = -3.03, R) >consensus CUCAAUUUUGUGACCCAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUUGUGUUGCACUUUGUCAACUCGUUUUUAAUGC ..(((..((.(((.((((((...(((((((((.....)))))))))))))))))).))..))).(((((.....).))))............. (-23.78 = -24.58 + 0.81)
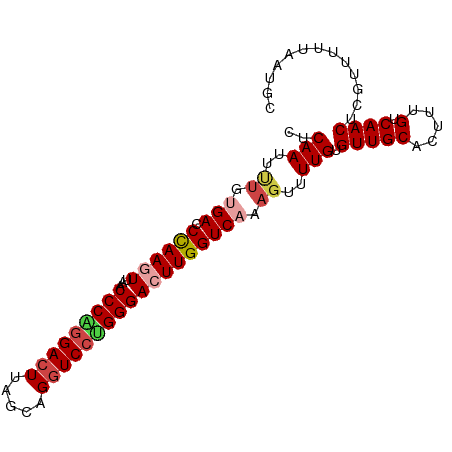
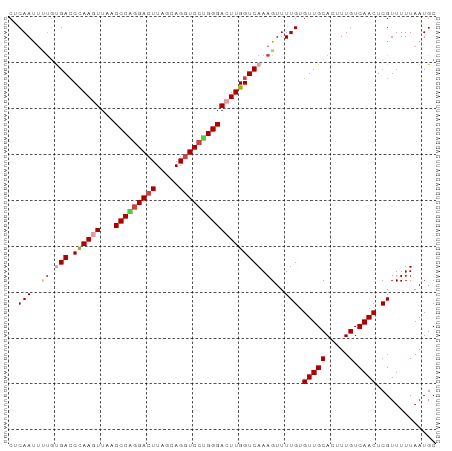
| Location | 6,040,914 – 6,041,009 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 86.34 |
| Shannon entropy | 0.25142 |
| G+C content | 0.48518 |
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -23.73 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6040914 95 + 24543557 AAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGGUCACAAAAUUGAGGGAA-GGGUGUUCCC---UUCAAACGUGAGUCACUCU- .......(((((((((((((((((.......))))))))...)))).))))).......(((.(((-(((....)))---)))..((....))..))).- ( -38.00, z-score = -3.84, R) >droAna3.scaffold_13337 16726028 92 + 23293914 AAAACUUUGACCAAGUCCCAGGACCUGCUAACUCUCGGGUUAACUUGGGUCGGGUAGUUGUGGGAG---GUGGUUCU-----CGAACGUGAGUCAUGUCA ........(((((..(((((.(((.((((....((((((....))))))...))))))).))))).---.)))))..-----.((.(....)))...... ( -31.50, z-score = -1.25, R) >droEre2.scaffold_4784 8738092 95 + 25762168 AAAACUUUGACCAAUUCCCGGGACCUGCUAAGUCCCGGGUUAACUUGGGUCGCAGAAUUGAGGGUA-GGGUGUUCCC---UUCAAACGUGAGUCACUCU- ....((.(((((....((((((((.......))))))))........))))).))....(((((.(-(((....)))---)))..((....))..))).- ( -33.60, z-score = -1.67, R) >droYak2.chr3L 6621944 99 + 24197627 AAAACUUUGACCAAUUCCCGGGACCUGCUAAGUCCUGGGUUAACUUGGGUCACAGCAUUGAGGAAAAGGGUGUUCCCCCCUUCAAACGUGAGUCACUCU- ....((.(((((....((((((((.......))))))))........))))).))..((((((....(((....)))..))))))((....))......- ( -31.40, z-score = -0.98, R) >droSec1.super_2 5990680 95 + 7591821 AAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGUUCACAAAAUUGAGGGAA-GGGUGCUCCC---UUCAAACGUGAGUCACUCU- .......((((((.((((((((((.......))))))))....(((((((.....))))))).(((-(((....)))---)))..)).)).))))....- ( -32.30, z-score = -2.49, R) >droSim1.chr3L 5543103 95 + 22553184 AAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGGUCACAAAAUUGAGGGAA-GGGUGCUCCC---UUCAAACGUGAGUCACUCU- .......(((((((((((((((((.......))))))))...)))).))))).......(((.(((-(((....)))---)))..((....))..))).- ( -38.00, z-score = -3.83, R) >consensus AAAACUUUGACCAAGUCCCAGGACCUGCUAAGUCCUGGGUUAACUUAGGUCACAAAAUUGAGGGAA_GGGUGUUCCC___UUCAAACGUGAGUCACUCU_ ...(((((((((((((((((((((.......))))))))...)))).))))).....(((((.....(((....)))...)))))....))))....... (-23.73 = -24.32 + 0.59)

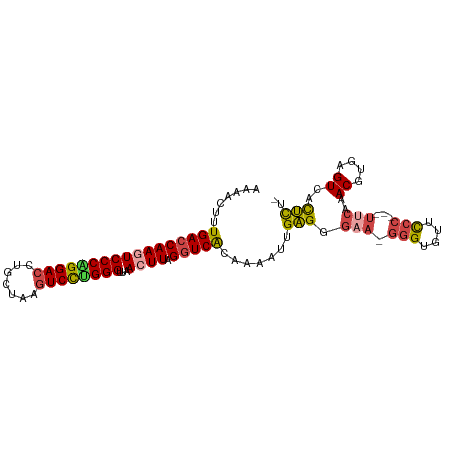
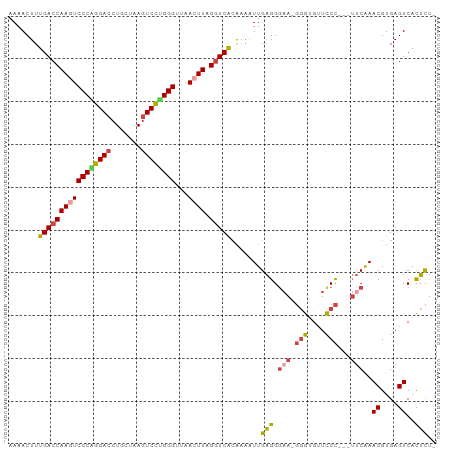
| Location | 6,040,914 – 6,041,009 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 86.34 |
| Shannon entropy | 0.25142 |
| G+C content | 0.48518 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -24.26 |
| Energy contribution | -25.65 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6040914 95 - 24543557 -AGAGUGACUCACGUUUGAA---GGGAACACCC-UUCCCUCAAUUUUGUGACCUAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUU -.(((.(((....))).(((---(((....)))-))).))).......(((((.((((...(((((((((.....))))))))))))))))))....... ( -36.20, z-score = -3.16, R) >droAna3.scaffold_13337 16726028 92 - 23293914 UGACAUGACUCACGUUCG-----AGAACCAC---CUCCCACAACUACCCGACCCAAGUUAACCCGAGAGUUAGCAGGUCCUGGGACUUGGUCAAAGUUUU ((((.((((((.((...(-----((......---)))....((((..........))))....)).)))))).((((((....))))))))))....... ( -21.60, z-score = -0.37, R) >droEre2.scaffold_4784 8738092 95 - 25762168 -AGAGUGACUCACGUUUGAA---GGGAACACCC-UACCCUCAAUUCUGCGACCCAAGUUAACCCGGGACUUAGCAGGUCCCGGGAAUUGGUCAAAGUUUU -.(((.(..(((....)))(---(((....)))-).).)))........((((..((((..(((((((((.....)))))))))))))))))........ ( -30.60, z-score = -1.37, R) >droYak2.chr3L 6621944 99 - 24197627 -AGAGUGACUCACGUUUGAAGGGGGGAACACCCUUUUCCUCAAUGCUGUGACCCAAGUUAACCCAGGACUUAGCAGGUCCCGGGAAUUGGUCAAAGUUUU -.(((.(((....))).((((((((.....)))))))))))...(((.(((((..((((..(((.(((((.....))))).)))))))))))).)))... ( -32.70, z-score = -0.91, R) >droSec1.super_2 5990680 95 - 7591821 -AGAGUGACUCACGUUUGAA---GGGAGCACCC-UUCCCUCAAUUUUGUGAACUAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUU -.....((((((((.((((.---(((((.....-)))))))))...)))))(((((((...(((((((((.....))))))))))))))))....))).. ( -36.30, z-score = -3.18, R) >droSim1.chr3L 5543103 95 - 22553184 -AGAGUGACUCACGUUUGAA---GGGAGCACCC-UUCCCUCAAUUUUGUGACCUAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUU -.(((.(((....))).(((---(((....)))-))).))).......(((((.((((...(((((((((.....))))))))))))))))))....... ( -36.20, z-score = -3.05, R) >consensus _AGAGUGACUCACGUUUGAA___GGGAACACCC_UUCCCUCAAUUUUGUGACCCAAGUUAACCCAGGACUUAGCAGGUCCUGGGACUUGGUCAAAGUUUU .....(((((((((.((((....(((((......)))))))))...)))))..(((((...(((((((((.....))))))))))))))))))....... (-24.26 = -25.65 + 1.39)
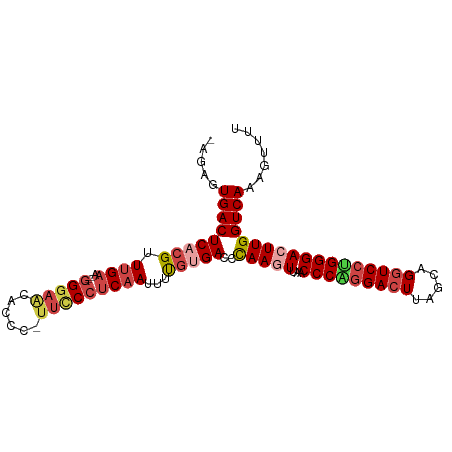
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:27 2011