| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,418,946 – 4,419,060 |
| Length | 114 |
| Max. P | 0.920786 |

| Location | 4,418,946 – 4,419,060 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.54159 |
| G+C content | 0.48585 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -23.16 |
| Energy contribution | -22.66 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4418946 114 + 23011544 ------------UAAUGU-UUUGCAAAAAAAAAACUUCUAAA-CUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACGUGAAAAAC ------------....((-(((........)))))(((....-..(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).....))).... ( -26.40, z-score = -0.92, R) >droSim1.chr2L 4343371 112 + 22036055 ------------UAAUGU-UGUGCAAAAAAAAC--UUCUGAU-CUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACGUGAAAAAC ------------......-.((((((.....((--(((.(((-(.((......((((.......))))......)).)))).)))))(((........)))))))))((((.......))))...... ( -26.50, z-score = -0.35, R) >droSec1.super_5 2517068 111 + 5866729 ------------UAAUGU-UUUGCAAAAAAAG---UUCCGAA-CUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACGUGAAAAAC ------------....((-(((.((.....((---(((((..-(((((((.((((.((.(((......)))...)))).)).)).)))))..((((.......))))..))))).))...)).))))) ( -27.90, z-score = -0.46, R) >droYak2.chr2L 4452561 115 + 22324452 ------------UAACGAAUUUGAAAACAAAAAAAUUCUUAA-CUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACGUGAAAAAU ------------..((((((((..........))))))....-..(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))...))....... ( -26.40, z-score = -1.19, R) >droEre2.scaffold_4929 4502768 112 + 26641161 ------------UAAGUA-CUCGGAAAAAAAAC--UUCUGAU-CUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACGUUAAAAAC ------------...(((-.((((((.......--)))))).-..(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))))......... ( -28.60, z-score = -0.91, R) >droAna3.scaffold_12943 1604785 114 - 5039921 ------------GAAAAUAAACAAAAAAAUAAUUAAUC--AACUUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCCACAUGCCAAUC ------------..........................--.....(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))............ ( -23.30, z-score = -0.73, R) >droPer1.super_8 3440795 128 - 3966273 CAAACAAAAGUAAAAGAAAAGCCAAAAAAUAAUGCUUCCAAAGUUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCCACAUAAGUCAC ...............((.((((...........)))).....((.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).)).....)).. ( -24.70, z-score = -0.27, R) >droWil1.scaffold_180772 4811408 125 - 8906247 -UAACGAUUUGUAAAAAAAAGGAUAUUUCCGAAGAAUCCAAU--UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCCACAUCCACCUC -....(((.((.........(((....)))............--.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))).)))...... ( -26.20, z-score = -0.39, R) >droMoj3.scaffold_6496 10232777 91 - 26866924 -----------------------AAAAAGGAAUUCUCUCAA--UUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUC------------ -----------------------..................--..(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))------------ ( -21.00, z-score = 0.11, R) >droVir3.scaffold_12855 5083947 102 - 10161210 -----------------------UAAAAAAAAUUAUUC-GAU-AUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAGUUGAAGCAA- -----------------------............(((-(((-.((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))))))))....- ( -27.90, z-score = -2.03, R) >droGri2.scaffold_14906 9431861 102 + 14172833 ------------------------AAAAAAUACUUUUCGAAA-UUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUC-ACUUGGAAUAC ------------------------..........((((((..-.((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))-).))))))... ( -25.70, z-score = -1.59, R) >anoGam1.chr2R 13972605 110 - 62725911 ----------------UAAAAAAAGUUAACUACGCAUUGCAU--UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCCGCAUGAAUGGC ----------------.............(((..(((.((..--.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).)))))..))). ( -26.70, z-score = -0.12, R) >consensus ____________UAA_GA_AUUGAAAAAAAAACUAUUCCAAA_CUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCUACGUGAAAAAC .............................................(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))............ (-23.16 = -22.66 + -0.50)

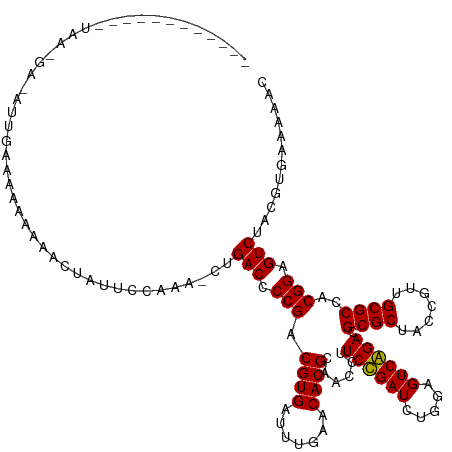
| Location | 4,418,946 – 4,419,060 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.54159 |
| G+C content | 0.48585 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -27.73 |
| Energy contribution | -27.23 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
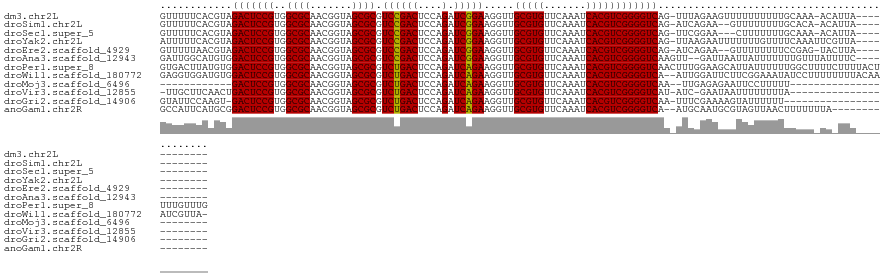
>dm3.chr2L 4418946 114 - 23011544 GUUUUUCACGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG-UUUAGAAGUUUUUUUUUUGCAAA-ACAUUA------------ (((((.((...(((((.(...((((.......))))....((((((.((((....))))(((((.......))))).))))))..-....).)))))......)).)))-))....------------ ( -31.70, z-score = -1.03, R) >droSim1.chr2L 4343371 112 - 22036055 GUUUUUCACGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG-AUCAGAA--GUUUUUUUUGCACA-ACAUUA------------ ((((.......))))...(((((((.......))))(((.((((((.((((....))))(((((.......))))).)))))).)-)).....--..........))).-......------------ ( -31.00, z-score = -0.51, R) >droSec1.super_5 2517068 111 - 5866729 GUUUUUCACGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG-UUCGGAA---CUUUUUUUGCAAA-ACAUUA------------ (((((.((...(((.((((..((((.......))))....((((((.((((....))))(((((.......))))).))))))..-..)))).---.)))...)).)))-))....------------ ( -33.60, z-score = -1.00, R) >droYak2.chr2L 4452561 115 - 22324452 AUUUUUCACGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG-UUAAGAAUUUUUUUGUUUUCAAAUUCGUUA------------ ......((((.......))))((((.......))))....((((((.((((....))))(((((.......))))).))))))..-....((((((..........))))))....------------ ( -31.20, z-score = -1.02, R) >droEre2.scaffold_4929 4502768 112 - 26641161 GUUUUUAACGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG-AUCAGAA--GUUUUUUUUCCGAG-UACUUA------------ .............((((....((((.......))))(((.((((((.((((....))))(((((.......))))).)))))).)-)).....--...........)))-).....------------ ( -31.40, z-score = -0.26, R) >droAna3.scaffold_12943 1604785 114 + 5039921 GAUUGGCAUGUGGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAAGUU--GAUUAAUUAUUUUUUUGUUUAUUUUC------------ (((..((....((((......((((.......))))))))((((((.((((....))))(((((.......))))).))))))..)).--.)))......................------------ ( -35.10, z-score = -1.58, R) >droPer1.super_8 3440795 128 + 3966273 GUGACUUAUGUGGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAACUUUGGAAGCAUUAUUUUUUGGCUUUUCUUUUACUUUUGUUUG ((((.....((.(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).))...((((((...........))))))...))))......... ( -31.10, z-score = 0.53, R) >droWil1.scaffold_180772 4811408 125 + 8906247 GAGGUGGAUGUGGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--AUUGGAUUCUUCGGAAAUAUCCUUUUUUUUACAAAUCGUUA- ((..((((((((....((((......))))...))))))(((((((.((((....))))(((((.......))))).)))))))--...((((((....))...)))).........))..))....- ( -36.90, z-score = -0.71, R) >droMoj3.scaffold_6496 10232777 91 + 26866924 ------------GACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAA--UUGAGAGAAUUCCUUUUU----------------------- ------------(((((((..((((.......)))).((((((....).))))).....(((((.......))))))))))))..--..(((((.....))))).----------------------- ( -27.40, z-score = -0.55, R) >droVir3.scaffold_12855 5083947 102 + 10161210 -UUGCUUCAACUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAU-AUC-GAAUAAUUUUUUUUA----------------------- -..........((((((((..((((.......)))).((((((....).))))).....(((((.......))))))))))))).-...-...............----------------------- ( -27.00, z-score = -0.56, R) >droGri2.scaffold_14906 9431861 102 - 14172833 GUAUUCCAAGU-GACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAA-UUUCGAAAAGUAUUUUUU------------------------ ...(((.((((-(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).)-))).)))...........------------------------ ( -28.70, z-score = -1.15, R) >anoGam1.chr2R 13972605 110 + 62725911 GCCAUUCAUGCGGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--AUGCAAUGCGUAGUUAACUUUUUUUA---------------- (((((((((...(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--))).)))).))...............---------------- ( -31.60, z-score = -0.12, R) >consensus GUUUUUCACGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG_UUUAGAAGAAUUUUUUUUUCAAA_UC_UUA____________ ............(((((((..((((.......)))).((((((....).))))).....(((((.......))))))))))))............................................. (-27.73 = -27.23 + -0.50)
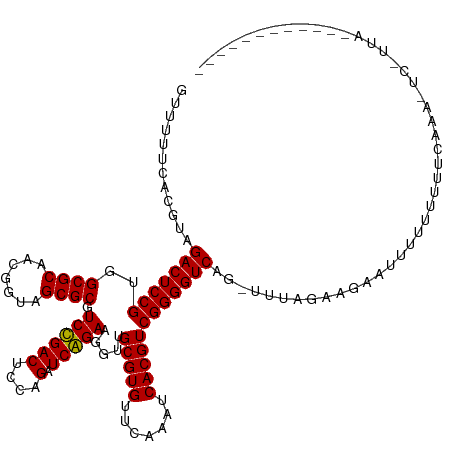
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:25 2011