
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,021,832 – 6,021,928 |
| Length | 96 |
| Max. P | 0.934442 |

| Location | 6,021,832 – 6,021,928 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Shannon entropy | 0.48361 |
| G+C content | 0.51388 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -15.14 |
| Energy contribution | -15.89 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
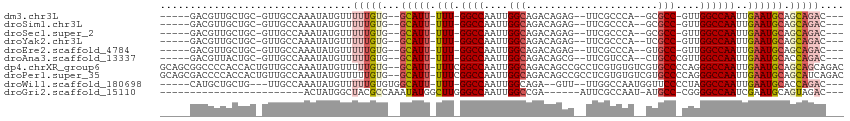
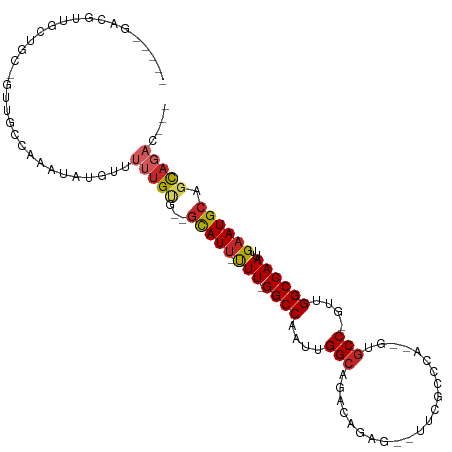
>dm3.chr3L 6021832 96 + 24543557 -----GACGUUGCUGC-GUUGCCAAAUAUGUUUUUGUG--GCAUU-UUU-GGCCAAUUGGCAGACAGAG--UUCGCCCA--GCGCC-GUUGGCCAAUUGAAUGCAGCAGAC--- -----....(((((((-(((((((((.(((((.....)--)))).-)))-)))((((((((.(((.(.(--((.....)--)).).-))).))))))))))))))))))..--- ( -40.10, z-score = -3.65, R) >droSim1.chr3L 5518506 96 + 22553184 -----GACGUUGCUGC-GUUGCCAAAUAUGUUUUUGUG--GCAUU-UUU-GGCCAAUUGGCAGACAGAG--UUCGCCCA--GCGCC-GUUGGCCAAUUGAAUGCAGCAGAC--- -----....(((((((-(((((((((.(((((.....)--)))).-)))-)))((((((((.(((.(.(--((.....)--)).).-))).))))))))))))))))))..--- ( -40.10, z-score = -3.65, R) >droSec1.super_2 5972483 96 + 7591821 -----GACGUUGCUGC-GUUGCCAAAUAUGUUUUUGUG--GCAUU-UUU-GGCCAAUUGGCAGACAGAG--UUCGCCCA--GCGCC-GUUGGCCAAUUGAAUGCAGCAGAC--- -----....(((((((-(((((((((.(((((.....)--)))).-)))-)))((((((((.(((.(.(--((.....)--)).).-))).))))))))))))))))))..--- ( -40.10, z-score = -3.65, R) >droYak2.chr3L 6603446 96 + 24197627 -----GACGUUGCUGC-GUUGCCAAAUAUGUUUUUGUG--GCAUU-UUU-GGCCAAUUGGCAGACAGAG--UUCGCCCA--UCGCC-GUUGGCCAAUUGAAUGCAGCAGAC--- -----....(((((((-(((((((((.(((((.....)--)))).-)))-)))((((((((.(((.(..--..)((...--..)).-))).))))))))))))))))))..--- ( -38.40, z-score = -3.43, R) >droEre2.scaffold_4784 8718065 96 + 25762168 -----GACGUUGCUGC-GUUGCCAAAUAUGUUUUUGUG--GCAUU-UUU-GGCCAAUUGGCAGACAGAG--UUCGCCCA--GUGCC-GUUGGCCAAUUGAAUGCAGCAGAC--- -----....(((((((-(((((((((.(((((.....)--)))).-)))-)))((((((((.(((.(.(--(.(.....--).)))-))).))))))))))))))))))..--- ( -38.90, z-score = -3.47, R) >droAna3.scaffold_13337 16709185 97 + 23293914 -----GACGUUACUGC-GUUGCCAAAUAUGUUUUUGUG--GCAUU-UUU-GGCCAAUUGGCAGACAGCG--UUCGUCCA--CUGCCCGUUGGCCAAUUGAAUGCACCAGAC--- -----(((((....))-)))............((((..--(((((-(((-(((((((.(((((...((.--...))...--))))).)))))))))..))))))..)))).--- ( -36.20, z-score = -3.17, R) >dp4.chrXR_group6 3659463 111 - 13314419 GCAGCGGCCCCACCACUGUUGCCAAAUAUGUUUUUGUG--GCAUU-UUUCGGCCAAUUGGCAGACAGCCGCCUCGUGUGUCGUGCCCCAGGGCCAAUUGAAUGCAGCAGCAGAC (((((((........))))))).......((((((((.--(((((-(...((((....((((((((..((...))..)))).))))....))))....)))))).)))).)))) ( -38.70, z-score = -0.68, R) >droPer1.super_35 69810 111 + 973955 GCAGCGACCCCACCACUGUUGCCAAAUAUGUUUUUGUG--GCAUU-UUUCGGCCAAUUGGCAGACAGCCGCCUCGUGUGUCGUGCCCCAGGGCCAAUUGAAUGCAGCAUCAGAC ((((((..........)))))).......((((.(((.--(((((-(...((((....((((((((..((...))..)))).))))....))))....)))))).)))..)))) ( -33.80, z-score = -0.28, R) >droWil1.scaffold_180698 3594982 97 + 11422946 -----CAUGCUGCUG---UUGCCAAAUAUGUUUUUGUGUGGCAUU-UUU-GGCCAAUUGGCAGA--GUU--UUGGCCAAUGGUUCCCCUAGGCCAAUUGAAUGCACCAGAC--- -----....(((.((---(.((((((.(((((.......))))).-)))-)))((((((((..(--(..--..((((...))))...))..))))))))...))).)))..--- ( -29.70, z-score = -0.58, R) >droGri2.scaffold_15110 13737419 79 - 24565398 ------------------------ACUAUGGCUACGCCAAAUAUGGCUUGGGCCAAUUGGCCGA------AUUCGCCAAU-AUGCC-CGGGGCCAAUCGAAUGCAGUAGAC--- ------------------------....((((..(((((....))))..((((..((((((.(.------...)))))))-..)))-))..))))................--- ( -28.30, z-score = -1.56, R) >consensus _____GACGUUGCUGC_GUUGCCAAAUAUGUUUUUGUG__GCAUU_UUU_GGCCAAUUGGCAGACAGAG__UUCGCCCA__GUGCC_GUUGGCCAAUUGAAUGCAGCAGAC___ ................................(((((...(((((.....((((....(((......................)))....)))).....))))).))))).... (-15.14 = -15.89 + 0.75)
| Location | 6,021,832 – 6,021,928 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.67 |
| Shannon entropy | 0.48361 |
| G+C content | 0.51388 |
| Mean single sequence MFE | -34.81 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6021832 96 - 24543557 ---GUCUGCUGCAUUCAAUUGGCCAAC-GGCGC--UGGGCGAA--CUCUGUCUGCCAAUUGGCC-AAA-AAUGC--CACAAAAACAUAUUUGGCAAC-GCAGCAACGUC----- ---((.((((((...((((((((..((-(((((--...)))..--..))))..))))))))(((-(((-.(((.--........))).))))))...-))))))))...----- ( -35.80, z-score = -3.42, R) >droSim1.chr3L 5518506 96 - 22553184 ---GUCUGCUGCAUUCAAUUGGCCAAC-GGCGC--UGGGCGAA--CUCUGUCUGCCAAUUGGCC-AAA-AAUGC--CACAAAAACAUAUUUGGCAAC-GCAGCAACGUC----- ---((.((((((...((((((((..((-(((((--...)))..--..))))..))))))))(((-(((-.(((.--........))).))))))...-))))))))...----- ( -35.80, z-score = -3.42, R) >droSec1.super_2 5972483 96 - 7591821 ---GUCUGCUGCAUUCAAUUGGCCAAC-GGCGC--UGGGCGAA--CUCUGUCUGCCAAUUGGCC-AAA-AAUGC--CACAAAAACAUAUUUGGCAAC-GCAGCAACGUC----- ---((.((((((...((((((((..((-(((((--...)))..--..))))..))))))))(((-(((-.(((.--........))).))))))...-))))))))...----- ( -35.80, z-score = -3.42, R) >droYak2.chr3L 6603446 96 - 24197627 ---GUCUGCUGCAUUCAAUUGGCCAAC-GGCGA--UGGGCGAA--CUCUGUCUGCCAAUUGGCC-AAA-AAUGC--CACAAAAACAUAUUUGGCAAC-GCAGCAACGUC----- ---((.((((((......((((((((.-(((((--((((....--.)))))).)))..))))))-)).-..(((--((............)))))..-))))))))...----- ( -37.00, z-score = -4.00, R) >droEre2.scaffold_4784 8718065 96 - 25762168 ---GUCUGCUGCAUUCAAUUGGCCAAC-GGCAC--UGGGCGAA--CUCUGUCUGCCAAUUGGCC-AAA-AAUGC--CACAAAAACAUAUUUGGCAAC-GCAGCAACGUC----- ---((.((((((......((((((((.-((((.--..((((..--...))))))))..))))))-)).-..(((--((............)))))..-))))))))...----- ( -35.40, z-score = -3.77, R) >droAna3.scaffold_13337 16709185 97 - 23293914 ---GUCUGGUGCAUUCAAUUGGCCAACGGGCAG--UGGACGAA--CGCUGUCUGCCAAUUGGCC-AAA-AAUGC--CACAAAAACAUAUUUGGCAAC-GCAGUAACGUC----- ---((.((.(((...((((((((....((((((--((......--))))))))))))))))(((-(((-.(((.--........))).))))))...-))).))))...----- ( -36.10, z-score = -3.15, R) >dp4.chrXR_group6 3659463 111 + 13314419 GUCUGCUGCUGCAUUCAAUUGGCCCUGGGGCACGACACACGAGGCGGCUGUCUGCCAAUUGGCCGAAA-AAUGC--CACAAAAACAUAUUUGGCAACAGUGGUGGGGCCGCUGC (..(((....)))..)..((((((....((((.((((..((...))..))))))))....))))))..-..(((--((............))))).((((((.....)))))). ( -38.50, z-score = -0.02, R) >droPer1.super_35 69810 111 - 973955 GUCUGAUGCUGCAUUCAAUUGGCCCUGGGGCACGACACACGAGGCGGCUGUCUGCCAAUUGGCCGAAA-AAUGC--CACAAAAACAUAUUUGGCAACAGUGGUGGGGUCGCUGC ....(((.(..(......((((((....((((.((((..((...))..))))))))....))))))..-..(((--((............)))))......)..).)))..... ( -34.80, z-score = 0.43, R) >droWil1.scaffold_180698 3594982 97 - 11422946 ---GUCUGGUGCAUUCAAUUGGCCUAGGGGAACCAUUGGCCAA--AAC--UCUGCCAAUUGGCC-AAA-AAUGCCACACAAAAACAUAUUUGGCAA---CAGCAGCAUG----- ---...((.(((......((((((..((....))((((((...--...--...)))))).))))-)).-..(((((..............))))).---..))).))..----- ( -29.74, z-score = -1.40, R) >droGri2.scaffold_15110 13737419 79 + 24565398 ---GUCUACUGCAUUCGAUUGGCCCCG-GGCAU-AUUGGCGAAU------UCGGCCAAUUGGCCCAAGCCAUAUUUGGCGUAGCCAUAGU------------------------ ---................((((..((-(((..-((((((....------...))))))..))))..((((....)))))..))))....------------------------ ( -29.20, z-score = -1.57, R) >consensus ___GUCUGCUGCAUUCAAUUGGCCAAC_GGCAC__UGGGCGAA__CUCUGUCUGCCAAUUGGCC_AAA_AAUGC__CACAAAAACAUAUUUGGCAAC_GCAGCAACGUC_____ ..........(((((.....((((....(((.....(((((.......))))))))....)))).....)))))........................................ (-14.15 = -14.57 + 0.42)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:24 2011