
| Sequence ID | dm3.chr3L |
|---|---|
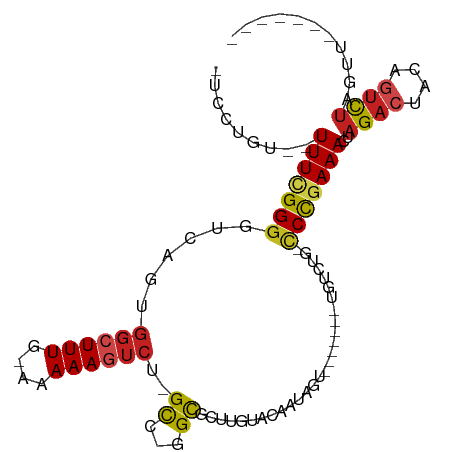
| Location | 6,016,501 – 6,016,588 |
| Length | 87 |
| Max. P | 0.802252 |

| Location | 6,016,501 – 6,016,588 |
|---|---|
| Length | 87 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.57335 |
| G+C content | 0.47494 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.88 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6016501 87 - 24543557 GUCCUGC--UUUCGGGGUCAGUGGCUUUG-AAAAAGUCU-GCU-GGCGCUUGUAGAAUAGU-------UGUCUG-CCCGAAACUAGACUACAGUCUAGUU------- .......--.(((((((((((..(.(((.-...))).).-.))-))).....((((.....-------..))))-))))))(((((((....))))))).------- ( -30.10, z-score = -2.48, R) >droSim1.chr3L 5513120 88 - 22553184 GUCCUGC-UUUUCGGGGUCAGUGGCUUUG-AAAAAGUCU-GCU-GGCGCUUGUAGAAUAGU-------UGUCUG-CCCGAAACUAGACUACAGUCUAGUU------- .......-..(((((((((((..(.(((.-...))).).-.))-))).....((((.....-------..))))-))))))(((((((....))))))).------- ( -30.10, z-score = -2.46, R) >droSec1.super_2 5967120 88 - 7591821 GUCCUGC-UUUUCGGGGUCAGUGGCUUUG-AAAAAGUCU-GCU-GGCGCUUGUAGAAUAGU-------UGUCCG-CCCGAAACUAGACUACAGUCUAGUU------- .......-..(((((((((((..(.(((.-...))).).-.))-)))(((........)))-------......-))))))(((((((....))))))).------- ( -29.70, z-score = -2.25, R) >droYak2.chr3L 6598001 90 - 24197627 GUCCUGUUUUUUCGGGGUCAGUGGCUUUG-AAAAAGUCU-GCC-GGCGCUUGUAGAAUAGU-------UGUCUGCCCCAAAACUAGACUGCAGUCUAGUU------- .............(((((..(..(((...-......(((-((.-.......)))))..)))-------..)..)))))..((((((((....))))))))------- ( -26.60, z-score = -1.50, R) >droEre2.scaffold_4784 8712626 87 - 25762168 GUCCUGU--UUUCGGGGUCAGUGGCUUUG-AAAAAGUCU-GCC-GGCGCUUGCAGAAUAGU-------UGUCUG-CCCGAAACUAGACUACAGUCUAGUU------- .......--.(((((.(((.(..(.(((.-...))).).-.).-)))....(((((.....-------..))))-))))))(((((((....))))))).------- ( -27.40, z-score = -1.48, R) >droAna3.scaffold_13337 16703784 86 - 23293914 GUCCUGU--UUUCGGGG--------UUUG-AAAAAGUCU-GCC-GGCGCUGGAACAAUAGU-------UGUCUG-CCCCAAAGUAGACUACGAACUAAAGUCUAACU ..((((.--...))))(--------((((-....(((((-((.-((.((.(((((....))-------).)).)-).))...))))))).)))))............ ( -20.50, z-score = -0.45, R) >dp4.chrXR_group6 3653914 95 + 13314419 --UCCGU--UUUCGGGGUCAGUGGCUUUGAAAAAAGUCU-GCCUGGCUCUUCUGCUACAAUGCUCUUGUGUCUGCCCCGAAACUAGACUACAGUCUAGGC------- --.....--((((((((.(((.((((((....)))))).-((.((((......))))....))........)))))))))))((((((....))))))..------- ( -34.70, z-score = -3.54, R) >droPer1.super_35 64268 95 - 973955 --UCCGU--UUUCGGGGUCAGUGGCUUUGAAAAAAGUCU-GCCUGGCUCUUCUGCUACAAUGCUCUUGUGUCUGCCCCGAAACUAGACUACAGUCUAGGC------- --.....--((((((((.(((.((((((....)))))).-((.((((......))))....))........)))))))))))((((((....))))))..------- ( -34.70, z-score = -3.54, R) >droWil1.scaffold_180698 7821387 87 + 11422946 ---UCGU-UUUUCGGGGUCAGUGGCUUUG-AAAAAGUCUGGCCCAGCUCUGGUACAAUGGA-------UGUCUA-GCUGAAAGUAGACUACAGUCUAGCU------- ---....-(((((.(((((((..((((..-...)))))))))))((((..((.(((.....-------))))))-))))))))(((((....)))))...------- ( -24.20, z-score = -0.42, R) >droVir3.scaffold_13049 3888809 87 - 25233164 ----CCUUUUUUUGGGGUCAGUGGCUUU--GAAAAGUCU-GUCUGGUUGUU-CACAAUGGC-------UGCG---UCUGAAAGUAGACUACAGUUUUGUGCGCUU-- ----(((......))).((((.(((((.--...))))).-..))))..((.-(((((.(((-------((.(---((((....)))))..)))))))))).))..-- ( -22.30, z-score = -0.40, R) >droMoj3.scaffold_6654 2351629 78 + 2564135 ---------UUUUGGGGUCAGUGGCUUU--GAAAA-UCU-GUCUGGCUGUA-UAGAACGGC-------UGCG---CCCGAAAGUAGACUACACUUUUGCGUU----- ---------(((..((((.....)))).--.))).-...-....((((((.-....)))))-------)(((---(..((((((.......)))))))))).----- ( -19.30, z-score = 0.16, R) >droGri2.scaffold_15110 13731597 88 + 24565398 -UUCUUUUUUUUUGGGGUCAGUGGCUUU--GAAAAGUCU-GUCUGGCUGUU-CACAAU-AC-------UA-G---UCUGAAACUAGACUACAGUUGUGCCAAGUG-- -.........(((((((((((.(((((.--...))))).-..))))))...-((((((-..-------((-(---((((....)))))))..)))))))))))..-- ( -27.50, z-score = -2.60, R) >consensus _UCCUGU__UUUCGGGGUCAGUGGCUUUG_AAAAAGUCU_GCC_GGCGCUUGUACAAUAGU_______UGUCUG_CCCGAAACUAGACUACAGUCUAGUU_______ .........(((((((......((((((....))))))..((...))............................)))))))..((((....))))........... (-10.88 = -10.88 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:22 2011