
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,004,344 – 6,004,435 |
| Length | 91 |
| Max. P | 0.971086 |

| Location | 6,004,344 – 6,004,435 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Shannon entropy | 0.49910 |
| G+C content | 0.53948 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -18.12 |
| Energy contribution | -19.23 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
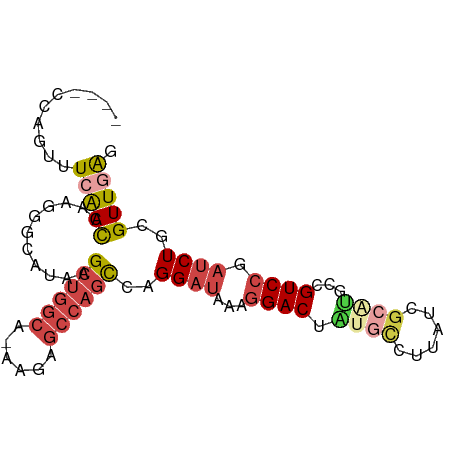
>dm3.chr3L 6004344 91 + 24543557 CCACCCAGUUUCAACCCACAGCACGAGCUGGCA-AAGAGCCAGCCAGGAUAAAGGACUAUGCCUUAUCGCAUGCCGUCCGAUCUGCGUUAGG ..............((....((..(.((((((.-....))))))).((((...(((((((((......)))))..)))).))))))....)) ( -27.40, z-score = -1.99, R) >droSim1.chr3L 5498522 91 + 22553184 CCACCCAGUUUCAACCAAGGGCAUGAGCUGGCA-AAGAGCCAGCCGGGAUAAAGGACUAUGCCUUAUCGCAUGCCGUCCGAUCUGCGUUGAG ..........((((((((.((((((.((((((.-....))))))...(((((.((......))))))).)))))).)......)).))))). ( -30.10, z-score = -1.39, R) >droSec1.super_2 5954887 91 + 7591821 CCACCCAGUUUCAACCAAGGGCAUGAGCUGGCA-AAGAGCCAGCCAGGAUAAAGGACUAUGCCUUAUCGCAUGCCGUCCGAUCUGCGUUGAG ..........((((((((.((((((.((((((.-....))))))...(((((.((......))))))).)))))).)......)).))))). ( -30.10, z-score = -1.86, R) >droEre2.scaffold_4784 8700458 71 + 25762168 ----UCAGUUUCGACAGUGGGCAUAAGCUGGCA-CAGAGCCAGCCAGGAUAAAGGACU----------------CGUCCGAUCUGCGUUGAG ----......(((((.(..(((....((((((.-....)))))).........(((..----------------..)))).))..)))))). ( -23.10, z-score = -1.48, R) >droYak2.chr3L 6584510 85 + 24197627 ------CGUUUAAAUGGAGGGCAUAAGCUGGCA-AAGAGCCAGCCAGGAUAAAGGACUGCGUCUUAUCGCAUUUCGUCCGAUCUGCGUUGAG ------......((((.((..(....((((((.-....))))))..((((...(((.((((......)))).))))))))..)).))))... ( -27.20, z-score = -1.48, R) >droAna3.scaffold_13337 16691290 88 + 23293914 ----GCAAUUUCAACUUUGGGAAUAAGCCCAAACAAGAGCCUUUAAGGCCGAACGACUCUGCCUUAUCGGGGCCUGUCCGAUCUGCGUUGAG ----......(((((((((((......))))))..((.(((((((((((.((.....)).))))))..)))))))...........))))). ( -27.00, z-score = -1.62, R) >consensus ____CCAGUUUCAACCAAGGGCAUAAGCUGGCA_AAGAGCCAGCCAGGAUAAAGGACUAUGCCUUAUCGCAUGCCGUCCGAUCUGCGUUGAG ..........(((((...........((((((......))))))..((((...((((.((((......))))...)))).))))..))))). (-18.12 = -19.23 + 1.11)


| Location | 6,004,344 – 6,004,435 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Shannon entropy | 0.49910 |
| G+C content | 0.53948 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -15.41 |
| Energy contribution | -17.58 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
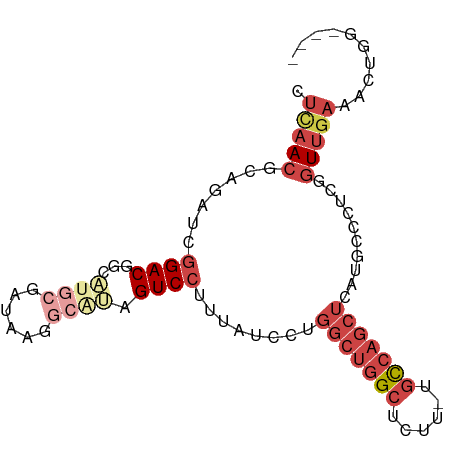
>dm3.chr3L 6004344 91 - 24543557 CCUAACGCAGAUCGGACGGCAUGCGAUAAGGCAUAGUCCUUUAUCCUGGCUGGCUCUU-UGCCAGCUCGUGCUGUGGGUUGAAACUGGGUGG ((((.....((((.(.(((((((.(((((((.......)))))))..(((((((....-.))))))))))))))).)))).....))))... ( -32.70, z-score = -1.13, R) >droSim1.chr3L 5498522 91 - 22553184 CUCAACGCAGAUCGGACGGCAUGCGAUAAGGCAUAGUCCUUUAUCCCGGCUGGCUCUU-UGCCAGCUCAUGCCCUUGGUUGAAACUGGGUGG ((((.....((((((..((((((.(((((((.......)))))))..(((((((....-.))))))))))))).)))))).....))))... ( -31.50, z-score = -0.98, R) >droSec1.super_2 5954887 91 - 7591821 CUCAACGCAGAUCGGACGGCAUGCGAUAAGGCAUAGUCCUUUAUCCUGGCUGGCUCUU-UGCCAGCUCAUGCCCUUGGUUGAAACUGGGUGG ((((.....((((((..((((((.(((((((.......)))))))..(((((((....-.))))))))))))).)))))).....))))... ( -31.40, z-score = -1.08, R) >droEre2.scaffold_4784 8700458 71 - 25762168 CUCAACGCAGAUCGGACG----------------AGUCCUUUAUCCUGGCUGGCUCUG-UGCCAGCUUAUGCCCACUGUCGAAACUGA---- .....(((((...(((..----------------..)))........(((((((....-.)))))))........))).)).......---- ( -20.50, z-score = -1.19, R) >droYak2.chr3L 6584510 85 - 24197627 CUCAACGCAGAUCGGACGAAAUGCGAUAAGACGCAGUCCUUUAUCCUGGCUGGCUCUU-UGCCAGCUUAUGCCCUCCAUUUAAACG------ ......(((....((((....((((......))))))))........(((((((....-.)))))))..)))..............------ ( -24.10, z-score = -2.22, R) >droAna3.scaffold_13337 16691290 88 - 23293914 CUCAACGCAGAUCGGACAGGCCCCGAUAAGGCAGAGUCGUUCGGCCUUAAAGGCUCUUGUUUGGGCUUAUUCCCAAAGUUGAAAUUGC---- .(((((.......(((((((..((..((((((.(((...))).))))))..))..)))))))(((......)))...)))))......---- ( -26.70, z-score = -0.92, R) >consensus CUCAACGCAGAUCGGACGGCAUGCGAUAAGGCAUAGUCCUUUAUCCUGGCUGGCUCUU_UGCCAGCUCAUGCCCUCGGUUGAAACUGG____ .(((((.......((((...((((......)))).))))........(((((((......)))))))..........))))).......... (-15.41 = -17.58 + 2.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:21 2011