
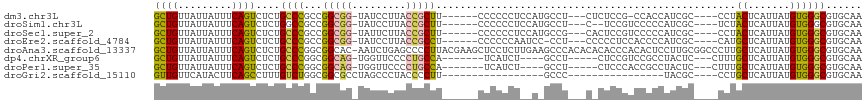
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,993,088 – 5,993,223 |
| Length | 135 |
| Max. P | 0.771953 |

| Location | 5,993,088 – 5,993,191 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Shannon entropy | 0.55631 |
| G+C content | 0.57596 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -11.24 |
| Energy contribution | -10.79 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5993088 103 + 24543557 GCUGUUAUUAUUUCAGUCUCUGCCCGCCGGCGG-UAUCCUUACCGCUU------CCCCCCUCCAUGCCU---CUCUCCG-CCACCAUCGC----CCUACUCAUUAUGUGGGCGUGCAA ((((.........))))...(((.....(((((-((....))))((..------...........))..---......)-)).....(((----((.((.......))))))).))). ( -23.32, z-score = -1.70, R) >droSim1.chr3L 5489892 102 + 22553184 GCUGUUAUUAUUUCAGUCUCUGGCCGCCGGCGG-UAUCCUUACCGCUU------CCCCCCUCCAUGCCU---C--UCCGUCCCCCAUCGC----UCUACUCAUUAUGUGGGCGUGCAA ((((.........)))).....(((((((((((-((....))))))).------...............---.--............(((----............))))))).)).. ( -22.90, z-score = -0.99, R) >droSec1.super_2 5946279 104 + 7591821 GCUGUUAUUAUUUCAGUCUCUGCCCGCCGGCGG-UAUUCUUACCGCUU------CCCCCCUCCAUGCCG---CACUCCGUCCCCCAUCGC----CCUACUCAUUAUGUGGGCGUGCAA ((((.........))))...........(((((-((....))))))).------..............(---(((...(........)((----((.((.......)))))))))).. ( -24.60, z-score = -1.68, R) >droEre2.scaffold_4784 8691823 103 + 25762168 GCUGUUAUUAUUUCAGUCUCUGCCCGCCGGCGG-UAUCCUUACCGCCU------CCCCCCAAUCC-CCU---CCCCCUCCACCCCAUCGC----CAUGCUCAUUAUGUGGGCGUGCAA ((((.........))))...(((.(((((((((-((....))))))).------...........-...---.......(((......((----...)).......))))))).))). ( -26.22, z-score = -3.53, R) >droAna3.scaffold_13337 16684036 117 + 23293914 GCUGUUAUUAUUUCAGUCUCUGCCCGGCGGCAC-AAUCUGAGCCCCUUACGAAGCUCCUCUUGAAGCCCACACACACCCACACUCCUUGCGGCCCUUGCUCAUUAUGUGGGCGUGCAA ..(((......(((((....((((....)))).-...)))))......)))..(((.(....).))).....(((.((((((.....((.(((....)))))...)))))).)))... ( -26.50, z-score = 0.76, R) >dp4.chrXR_group6 3630349 98 - 13314419 GCUGUUAUUAUUUCAGUCUCUGCCCGGCGGCAG-UGGUUCCCCUGCCA-------UCAUCU----GCCU-----CUCCGUCCGCCUACUC---CUUUGCUCAUUAUGUGGGCGUGCAA ((((.........))))........(((((..(-((((......))))-------)...))----))).-----....(..(((((((..---.............)))))))..).. ( -28.16, z-score = -1.93, R) >droPer1.super_35 41686 98 + 973955 GCUGUUAUUAUUUCAGUCUCUGCCCGGCGGCAG-UGGUUCCCCUGCCA-------UCAUCU----GCCU-----CUCCCACCGCCUACUC---CUUUGCUCAUUAUGUGGGCGUGCAA ((((.........))))...(((..(((((..(-((((......))))-------)...))----))).-----.......(((((((..---.............))))))).))). ( -27.26, z-score = -1.58, R) >droGri2.scaffold_15110 13709963 81 - 24565398 GUUGUUCAUACUUCAGCCUUUGUCUGGCGGCGCCUAGCCCUACCCCUU-----------------GCCC----------------UACGC----CCUGCUCAUUAUGUGGGCGUGCAA ...............(((.......)))(((.....)))........(-----------------((..----------------.((((----((.((.......))))))))))). ( -22.20, z-score = -0.41, R) >consensus GCUGUUAUUAUUUCAGUCUCUGCCCGCCGGCGG_UAUCCUUACCGCUU______CCCCCCU__A_GCCU___C_CUCCGUCCCCCAUCGC____CCUGCUCAUUAUGUGGGCGUGCAA ((((.........))))....((((...(((((.........)))))..................................................((.......))))))...... (-11.24 = -10.79 + -0.45)


| Location | 5,993,126 – 5,993,223 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.78 |
| Shannon entropy | 0.52954 |
| G+C content | 0.50322 |
| Mean single sequence MFE | -15.36 |
| Consensus MFE | -9.16 |
| Energy contribution | -8.74 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5993126 97 + 24543557 ------------UUACCGCUUCCCCCCUCCAUGCCUCUCUCCG-CCACCAUCGCCCU-ACUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ------------..................((((.........-.......(((((.-((.......))))))).((.....)).))))...................... ( -13.80, z-score = -1.20, R) >droSim1.chr3L 5489930 96 + 22553184 ------------UUACCGCUUCCCCCCUCCAUGCCUC--UCCGUCCCCCAUCGCUCU-ACUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCCUUUACCGUUAUCUCUUU ------------..................((((...--............(((.((-((.......))))))).((.....)).))))...................... ( -11.50, z-score = -0.81, R) >droSec1.super_2 5946317 98 + 7591821 ------------UUACCGCUUCCCCCCUCCAUGCCGCACUCCGUCCCCCAUCGCCCU-ACUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ------------..................((((.(((..........((.(((((.-((.......))))))))).....))).))))...................... ( -15.56, z-score = -1.48, R) >droYak2.chr3L 6575802 97 + 24197627 ------------UUACCGCUUCCCCCCCAU-UGCCUCCCUCUUUGCCCCAUCGCCCU-ACUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ------------..................-(((....(..(((((.....(((((.-((.......))))))).)))))..)..)))....................... ( -14.90, z-score = -1.74, R) >droAna3.scaffold_13337 16684074 111 + 23293914 GAGCCCCUUACGAAGCUCCUCUUGAAGCCCACACACACCCACACUCCUUGCGGCCCUUGCUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ((((..(....)..))))....(((((((((((............((....)).............)))))))(((((......))))).))))................. ( -22.51, z-score = -0.82, R) >dp4.chrXR_group6 3630387 92 - 13314419 ---------------CCCCUGCCAUCAUCUGCCUCUCCGUCCG---CCUACUCCUUU-GCUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ---------------...........................(---(((((......-.........))))))(((((......)))))...................... ( -13.66, z-score = -0.79, R) >droPer1.super_35 41724 92 + 973955 ---------------CCCCUGCCAUCAUCUGCCUCUCCCACCG---CCUACUCCUUU-GCUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ---------------...........................(---(((((......-.........))))))(((((......)))))...................... ( -13.66, z-score = -0.90, R) >droGri2.scaffold_15110 13709994 82 - 24565398 ---------------------------GCCUAGCCCUACCCCUUGCCCUA-CGCCCU-GCUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUAGCA ---------------------------...............((((...(-(((((.-((.......))))))))))))..(((((.(((............))).))))) ( -17.30, z-score = -0.52, R) >consensus ____________UUACCGCUUCCCCCCUCCAUGCCUCCCUCCGUCCCCCAUCGCCCU_ACUCAUUAUGUGGGCGUGCAAAUUGCUGCAUAUUCAUUUACCGUUAUCUCUUU ....................................................((((..((.......))))))(((((......)))))...................... ( -9.16 = -8.74 + -0.42)

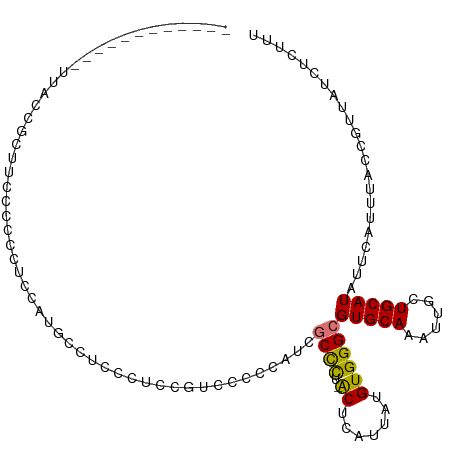
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:20 2011