
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,956,211 – 5,956,315 |
| Length | 104 |
| Max. P | 0.500000 |

| Location | 5,956,211 – 5,956,315 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.55755 |
| G+C content | 0.45880 |
| Mean single sequence MFE | -14.13 |
| Consensus MFE | -7.89 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
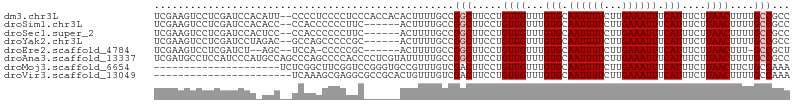
>dm3.chr3L 5956211 104 + 24543557 UCGAAGUCCUCGAUCCACAUU--CCCCUCCCCUCCCACCACACUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCGCC ((((.....))))........--..........................((((.....((((...(((.((((((...)))))).)))....))))....)))).. ( -13.20, z-score = -1.39, R) >droSim1.chr3L 5451470 98 + 22553184 UCGAAGUCCUCGAUCCACACC--CCACCCCCCUUC------ACUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCGCC ((((.....))))........--............------........((((.....((((...(((.((((((...)))))).)))....))))....)))).. ( -13.20, z-score = -1.45, R) >droSec1.super_2 5909551 98 + 7591821 UCGAAGUCCUCGAUCCACUCC--CCACCCCCCUUC------ACUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCGCC ((((.....))))........--............------........((((.....((((...(((.((((((...)))))).)))....))))....)))).. ( -13.20, z-score = -1.60, R) >droYak2.chr3L 6536489 98 + 24197627 UCGAAGUCCUCGAUCCUAGAC--GCCAGCCCCCGC------ACUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCGCC ((((.....))))...(((((--(.(((..((.((------(....))).))....))).)))))).((((.......(((((...))))).......)))).... ( -16.14, z-score = -0.42, R) >droEre2.scaffold_4784 8653237 94 + 25762168 UCGAAGUCCUCGAUCU--AGC--UCCA-CCCCCGC------ACUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUU-GCCGCU ((((.....))))...--(((--....-.....((------(....))).(((.....((((...(((.((((((...)))))).)))....))))...-)))))) ( -15.00, z-score = -0.89, R) >droAna3.scaffold_13337 16646487 106 + 23293914 UCGAUGCCUCCAUCCCAUGCCAGCCCAGCCCCACCCCUCGUAUUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCGCC ..((((....)))).........................(((....)))((((.....((((...(((.((((((...)))))).)))....))))....)))).. ( -12.90, z-score = 0.13, R) >droMoj3.scaffold_6654 2287707 85 - 2564135 ---------------------UCUCGGCUUCGGUCCGGGUGCCGUUUGUCGACUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUCUGCCAAA ---------------------..(((((..((((......))))...)))))..........((((.(((........(((((...)))))........))))))) ( -15.49, z-score = -0.51, R) >droVir3.scaffold_13049 3802250 83 + 25233164 -----------------------UCAAAGCGAGGCGCCGCACUGUUUGUCGACUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCAAA -----------------------.........(((...((((.......((((.....))))...)))).........(((((...))))).........)))... ( -13.90, z-score = 0.27, R) >consensus UCGAAGUCCUCGAUCCA_ACC__CCCACCCCCUGC______ACUUUUGCCGGCUUCCUGUUGUUUGUGCAAUUUUCUUGAAAUUUCAUUUCUUAACUUUUGCCGCC ..................................................(((.....((((...(((.((((((...)))))).)))....))))....)))... ( -7.89 = -8.14 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:16 2011