
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,948,387 – 5,948,456 |
| Length | 69 |
| Max. P | 0.708498 |

| Location | 5,948,387 – 5,948,456 |
|---|---|
| Length | 69 |
| Sequences | 11 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Shannon entropy | 0.17105 |
| G+C content | 0.43534 |
| Mean single sequence MFE | -17.36 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
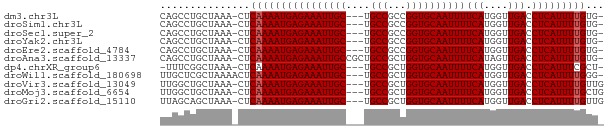
>dm3.chr3L 5948387 69 + 24543557 CAGCCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCCGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUG- ............-(.((((((((((((((((---.(((...))))))))))(((....))).))))))))).)- ( -17.30, z-score = -1.54, R) >droSim1.chr3L 5443355 69 + 22553184 CAGCCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCCGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUG- ............-(.((((((((((((((((---.(((...))))))))))(((....))).))))))))).)- ( -17.30, z-score = -1.54, R) >droSec1.super_2 5901764 69 + 7591821 CAGCCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCCGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUG- ............-(.((((((((((((((((---.(((...))))))))))(((....))).))))))))).)- ( -17.30, z-score = -1.54, R) >droYak2.chr3L 6528355 69 + 24197627 CAGCCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCCGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUG- ............-(.((((((((((((((((---.(((...))))))))))(((....))).))))))))).)- ( -17.30, z-score = -1.54, R) >droEre2.scaffold_4784 8645640 69 + 25762168 CAGCCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCCGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUG- ............-(.((((((((((((((((---.(((...))))))))))(((....))).))))))))).)- ( -17.30, z-score = -1.54, R) >droAna3.scaffold_13337 16638179 72 + 23293914 CAGCCUGCUAAA-CUCAAAAUGAGAAAUUGCCGCUGCCGCUGGUGCAAUUUUCAUAGUUGACCUCAUUUUGUG- ............-(.((((((((((((((((.(((......))))))))))(((....))).))))))))).)- ( -17.10, z-score = -1.37, R) >dp4.chrXR_group6 3572566 68 - 13314419 -UUUCGGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCUGGUGCAAUUUUCAUGGUUGACCUCAUUUCGCU- -..(((((((..-(((.....)))(((((((---.(((...))))))))))...)))))))............- ( -15.40, z-score = -0.89, R) >droWil1.scaffold_180698 3696592 70 + 11422946 UUGCUCGCUAAAACUCAAAAUGAGAAAUUGC---UGCCGCUGGUGCAAUUUUCAUGGUUGACCUCAUUUUGGG- .............((((((((((((((((((---.(((...))))))))))(((....))).)))))))))))- ( -20.60, z-score = -2.63, R) >droVir3.scaffold_13049 3785515 70 + 25233164 UUGGCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCUGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUUG ............-..((((((((((((((((---.(((...))))))))))(((....))).)))))))))... ( -17.20, z-score = -1.45, R) >droMoj3.scaffold_6654 2278111 70 - 2564135 UUGGCUGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCUGGUGCAAUUUUCAUGGUUGACCUCAUUUUGCUG ............-..((((((((((((((((---.(((...))))))))))(((....))).)))))))))... ( -17.00, z-score = -1.25, R) >droGri2.scaffold_15110 13661360 70 - 24565398 UUAGCAGCUAAA-CUCAAAAUGAGAAAUUGC---UGCCGCUGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUUG ............-..((((((((((((((((---.(((...))))))))))(((....))).)))))))))... ( -17.20, z-score = -1.29, R) >consensus CAGCCUGCUAAA_CUCAAAAUGAGAAAUUGC___UGCCGCUGGUGCAAUUUUCAUGGUUGACCUCAUUUUGUG_ ...............((((((((((((((((....(((...))))))))))(((....))).)))))))))... (-15.16 = -15.25 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:16 2011