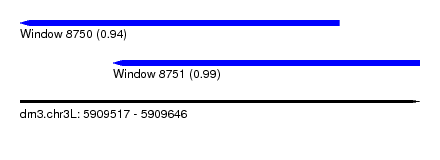
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,909,517 – 5,909,646 |
| Length | 129 |
| Max. P | 0.994499 |

| Location | 5,909,517 – 5,909,620 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.14 |
| Shannon entropy | 0.50523 |
| G+C content | 0.47246 |
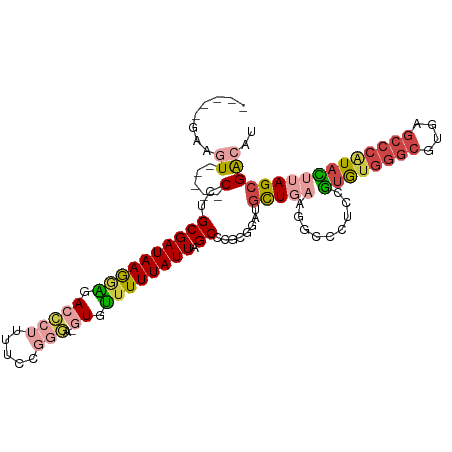
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
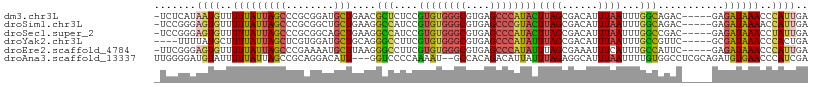
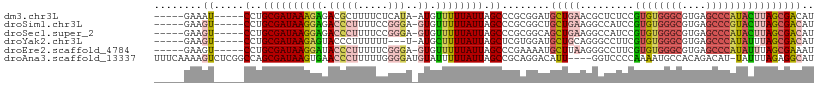
>dm3.chr3L 5909517 103 - 24543557 -UCUCAUAAUGUUUUUAUUAGCCCGCGGAUGCUGAACGCUCUCCGUGUGGGCGUGAGCCCAUACUUAGCGACAUUUAAUUUGGCAGAC-----GAGAUAAACCCAUUGA -.....(((((..((((((......((..(((..((((((....((((((((....))))))))..))))........))..)))..)-----).))))))..))))). ( -35.00, z-score = -3.64, R) >droSim1.chr3L 5402581 103 - 22553184 -UCCGGGAGUGUUUUUAUUAGCCCGCGGCUGCUGAAGGCCAUCCGUGUGGGCGUGAGCCCGUACUUAGCGACAUUUAAUUUGGCAGAC-----GAGAUAAAACCAUUGA -...((...((((((.....((((((((((......))))....((((((((....))))))))...)))...........)))....-----))))))...))..... ( -32.20, z-score = -0.87, R) >droSec1.super_2 5845184 103 - 7591821 -UCCGGGAGUGUUUUUAUUAGCCCGCGGCAGCUGAAGGCCAUCCGUGUGGGCGUGAGCCCAUACUUAGCGACAUUUAAUUUGGCCGAC-----GAGAUAAACCUAUUGA -..((((...((....))...))))((((.(((((.((....))((((((((....)))))))))))))..((.......))))))..-----................ ( -35.40, z-score = -2.03, R) >droYak2.chr3L 6486962 100 - 24197627 ----UUUUAUGCUUUUAUUAGCUCGUGGAUGCUGCAGGGCCUUCGUGUGGGCGUGAGCCCAUAUUUAGCGACAUUUAAUUUGCCGUUC-----GCGAUAAACCCACUGA ----......(((......)))(((((((((..(((((.(((..((((((((....))))))))..)).)........))))))))))-----))))............ ( -31.70, z-score = -1.78, R) >droEre2.scaffold_4784 8606589 103 - 25762168 -UUCGGGAGUGUUUUUAUUAGCCCGAAAAUGCUUAAGGGCCUUCGUGUGGGCGUGAGCCCAUAUUUAGCGAAAUUUCAUUUGCCAUUC-----GAGAUAAACCCAUUGA -...(((..((((((.....((((............))))....((((((((....))))))))...(((((......))))).....-----))))))..)))..... ( -33.00, z-score = -2.53, R) >droAna3.scaffold_13337 10688511 104 + 23293914 UUGGGGAUGUAUUUUUAUUAGCCGCAGGACAUU---GGUCCCCAAAAU--GCCACAGACAUUAUUUAGAGGCAUUUAAUUUUGUGGCCUCGCAGAUGUGAACCCAUCGA .((((...............((((((((((...---.))))...((((--(((.(.((......)).).))))))).....)))))).((((....)))).)))).... ( -30.20, z-score = -1.80, R) >consensus _UCCGGGAGUGUUUUUAUUAGCCCGCGGAUGCUGAAGGCCCUCCGUGUGGGCGUGAGCCCAUACUUAGCGACAUUUAAUUUGGCAGAC_____GAGAUAAACCCAUUGA .......((((..(((((((((........)))....(((....((((((((....))))))))((((......))))...)))...........))))))..)))).. (-15.61 = -15.73 + 0.12)
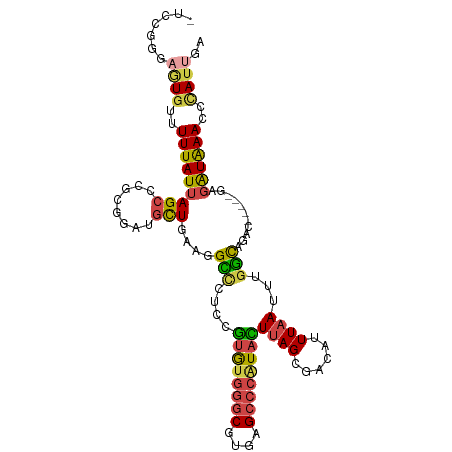
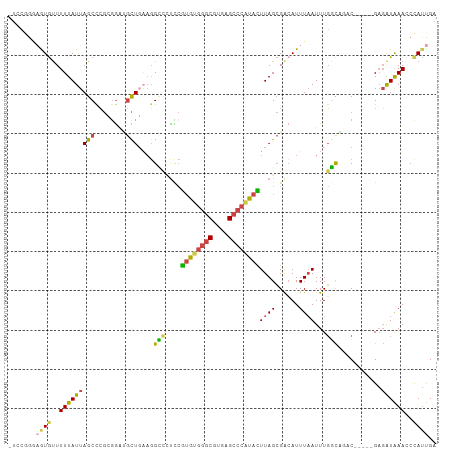
| Location | 5,909,547 – 5,909,646 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.44 |
| Shannon entropy | 0.47957 |
| G+C content | 0.49641 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -17.03 |
| Energy contribution | -18.65 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5909547 99 - 24543557 -----GAAAU-----CCUGCGAUAAAGAGACGCUUUUCUCAUA-AUGUUUUUAUUAGCCCGCGGAUGCUGAACGCUCUCCGUGUGGGCGUGAGCCCAUACUUAGCGACAU -----...((-----((.(((.....((((......)))).((-(((....)))))...)))))))......((((....((((((((....))))))))..)))).... ( -34.60, z-score = -3.44, R) >droSim1.chr3L 5402611 99 - 22553184 -----GAAGU-----CCUGCGAUAAGGAGACCCUUUUCCGGGA-GUGUUUUUAUUAGCCCGCGGCUGCUGAAGGCCAUCCGUGUGGGCGUGAGCCCGUACUUAGCGACAU -----..(.(-----((((.((.((((....)))).)))))))-.).............(((((((......))))....((((((((....))))))))...))).... ( -37.80, z-score = -2.18, R) >droSec1.super_2 5845214 99 - 7591821 -----GAAGU-----CCUGCGAUAAGGAGACCCUUUUCCGGGA-GUGUUUUUAUUAGCCCGCGGCAGCUGAAGGCCAUCCGUGUGGGCGUGAGCCCAUACUUAGCGACAU -----....(-----((((.((.((((....)))).)))))))-(((((.......(((...))).(((((.((....))((((((((....)))))))))))))))))) ( -40.70, z-score = -3.36, R) >droYak2.chr3L 6486992 96 - 24197627 -----GAAGU-----CCUGCGAUAAGAGUACCCUUUUUU---U-AUGCUUUUAUUAGCUCGUGGAUGCUGCAGGGCCUUCGUGUGGGCGUGAGCCCAUAUUUAGCGACAU -----(((((-----(((((((((((((((.........---.-.))))))))))(((........))))))))).))))((((((((....)))))))).......... ( -39.10, z-score = -4.39, R) >droEre2.scaffold_4784 8606619 99 - 25762168 -----GAAGU-----CCUGCGAUAAGGAUACCCUUUUUCGGGA-GUGUUUUUAUUAGCCCGAAAAUGCUUAAGGGCCUUCGUGUGGGCGUGAGCCCAUAUUUAGCGAAAU -----(((((-----(((......))))..((((((((((((.-..((....))...)))))))......))))).))))((((((((....)))))))).......... ( -36.90, z-score = -3.55, R) >droAna3.scaffold_13337 10688546 105 + 23293914 UUUCAAAAGUCUCGGCCAGCGAUAAGUGAACCCUUUUUGGGGAUGUAUUUUUAUUAGCCGCAGGACAUU----GGUCCCCAAAAUGCCACAGACAU-UAUUUAGAGGCAU ........((((((((.((..(.(((((..((((....))))...))))))..)).)))(((((((...----.))))......))).........-......))))).. ( -24.60, z-score = -0.46, R) >consensus _____GAAGU_____CCUGCGAUAAGGAGACCCUUUUCCGGGA_GUGUUUUUAUUAGCCCGCGGAUGCUGAAGGCCCUCCGUGUGGGCGUGAGCCCAUACUUAGCGACAU ..................((((((((((.(((((.....)))..)).)))))))).)).......((((((.........((((((((....)))))))))))))).... (-17.03 = -18.65 + 1.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:15 2011