
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,869,399 – 5,869,533 |
| Length | 134 |
| Max. P | 0.970644 |

| Location | 5,869,399 – 5,869,503 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.96 |
| Shannon entropy | 0.55451 |
| G+C content | 0.37121 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -10.05 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
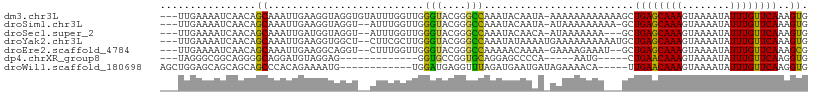
>dm3.chr3L 5869399 104 + 24543557 ---UUGAAAAUCAACAGCAAAUUGAAGGUAGGUGUAUUUGGUUGGGUACGGGCCAAAUACAAUA-AAAAAAAAAAAAGCUGAGCAAAGUAAAAUAUUUGUUCAAAGUG ---............(((..............(((((((((((.(...).)))))))))))...-............)))(((((((........)))))))...... ( -21.05, z-score = -2.63, R) >droSim1.chr3L 5362658 101 + 22553184 ---UUGAAAAUCAACAGCAAAUUGAAGGUAGGU--AUUUGGUUGGGUACGGGCCAAAUACAAUA-AUAAAAAAAAA-GCUGAGCAAAGUAAAAUAUUUGUUCAAAGUG ---.............((.............((--((((((((.(...).))))))))))....-...........-..((((((((........))))))))..)). ( -19.50, z-score = -2.29, R) >droSec1.super_2 5805245 99 + 7591821 ---UUGAAAAUCAACAGCAAAUUGAUGGUAGGU--AUUUGGUUGGGUACGGGCCAAAUACAACA-AUAAAAAAA---GCUGAGCAAAGUAAAAUAUUUGUUCAAAGUG ---(((...((((.(((....))).))))..((--((((((((.(...).))))))))))..))-)........---..((((((((........))))))))..... ( -20.90, z-score = -2.21, R) >droYak2.chr3L 6445252 103 + 24197627 ---UUGAAAAUCAACAGCAAAUUGAAGGUGGCU--CUUCGCUUGGGUACGGGCCAAAUAUAAAAUGAAAAAAAAAAUGCUGAGCAAAGUAAAAUAUUUGUUCAAAGUG ---............((((....(((((....)--))))(((((....))))).......................))))(((((((........)))))))...... ( -19.30, z-score = -0.98, R) >droEre2.scaffold_4784 8564768 100 + 25762168 ---UUGAAAAUCAACAGCAAAUUGAAGGCAGGU--CUUUGGUUGGGUACGGGCCAAAAACAAAA-GAAAAGAAAU--GCUGAGCAAAGUAAAAUAUUUGUUCAAAGCG ---.............((........((((..(--((((.(((.(((....)))...)))....-..)))))..)--)))(((((((........)))))))...)). ( -22.80, z-score = -2.14, R) >dp4.chrXR_group8 5707362 82 - 9212921 ---UAGGGCGGCAGGGGCAGGAUGUAGGAG-------------GGUGCCGGUGCAGGAGCCCCA-----AAUG-----CUGAACAAAGUAAAAUAUUUGUUCAAGGUG ---...((((...(((((....((((((..-------------....))..))))...))))).-----..))-----))(((((((........)))))))...... ( -25.00, z-score = -1.41, R) >droWil1.scaffold_180698 2128204 91 - 11422946 AGCUGGAGCAGCAGCAGCCCACAGAAAAUG------------UGGAUGAGGUUUAGAUGAAUGAUAGAAAACA-----UUGAACAAAGUAAAAUAUUUGUUCAAGGUG .((((......))))...(((((.....))------------))).........................((.-----(((((((((........))))))))).)). ( -21.60, z-score = -2.48, R) >consensus ___UUGAAAAUCAACAGCAAAUUGAAGGUAGGU__AUUUGGUUGGGUACGGGCCAAAUACAAAA_AAAAAAAAA___GCUGAGCAAAGUAAAAUAUUUGUUCAAAGUG ................((..........................(((....))).........................((((((((........))))))))..)). (-10.05 = -9.10 + -0.95)


| Location | 5,869,426 – 5,869,533 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.89 |
| Shannon entropy | 0.55280 |
| G+C content | 0.38698 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -10.13 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
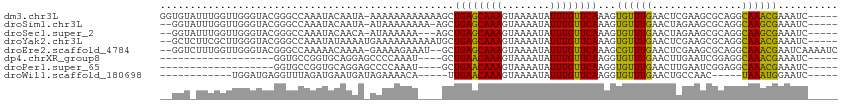
>dm3.chr3L 5869426 107 + 24543557 GGUGUAUUUGGUUGGGUACGGGCCAAAUACAAUA-AAAAAAAAAAAAGCUGAGCAAAGUAAAAUAUUUGUUCAAAGUGUUUGAACUCGAAGCGCAGGCAAACGAAAUC----- ..(((((((((((.(...).)))))))))))...-............((((((((((........)))))))...((((((.......)))))).)))..........----- ( -27.20, z-score = -3.45, R) >droSim1.chr3L 5362685 104 + 22553184 --GGUAUUUGGUUGGGUACGGGCCAAAUACAAUA-AUAAAAAAAA-AGCUGAGCAAAGUAAAAUAUUUGUUCAAAGUGUUUGAACUAGAAGCGCAGGCAAGCGAAAUC----- --.((((((((((.(...).))))))))))....-..........-.((((((((((........)))))))...((((((.......)))))).)))..........----- ( -26.30, z-score = -3.37, R) >droSec1.super_2 5805272 102 + 7591821 --GGUAUUUGGUUGGGUACGGGCCAAAUACAACA-AUAAAAAA---AGCUGAGCAAAGUAAAAUAUUUGUUCAAAGUGUUUGAACUAGAAGCGCAGGCAAGCGAAAUC----- --.((((((((((.(...).))))))))))....-........---.((((((((((........)))))))...((((((.......)))))).)))..........----- ( -26.30, z-score = -3.03, R) >droYak2.chr3L 6445279 106 + 24197627 --GCUCUUCGCUUGGGUACGGGCCAAAUAUAAAAUGAAAAAAAAAAUGCUGAGCAAAGUAAAAUAUUUGUUCAAAGUGUUUGAACUCGAAGCGCAGGCAAACGAAAUC----- --((.((.(((((((((.((((((..........(....).........((((((((........))))))))..).))))).)))).))))).))))..........----- ( -24.40, z-score = -1.75, R) >droEre2.scaffold_4784 8564795 108 + 25762168 --GGUCUUUGGUUGGGUACGGGCCAAAAACAAAA-GAAAAGAAAU--GCUGAGCAAAGUAAAAUAUUUGUUCAAAGCGUUUGAACUCGAAGCGCAGGCAAACGAAUCAAAAUC --..(((((.(((.(((....)))...))).)))-)).......(--((((((((((........)))))))...((((((.......)))))).)))).............. ( -25.60, z-score = -1.81, R) >dp4.chrXR_group8 5707389 85 - 9212921 -------------------GGUGCCGGUGCAGGAGCCCCAAAU----GCUGAACAAAGUAAAAUAUUUGUUCAAGGUGUUUGAACUUGAAUCGGAGGCAAACGAAAUC----- -------------------.((.(((((.((((.....(((((----((((((((((........)))))))..))))))))..)))).)))))..))..........----- ( -22.40, z-score = -1.38, R) >droPer1.super_65 89596 85 - 397436 -------------------GGUGCCGGUGCAGGAGCCCCAAAU----GCUGAACAAAGUAAAAUAUUUGUUCAAGGUGUUUGAACUUGAAUCGGAGGCAAACGAAAUC----- -------------------.((.(((((.((((.....(((((----((((((((((........)))))))..))))))))..)))).)))))..))..........----- ( -22.40, z-score = -1.38, R) >droWil1.scaffold_180698 2128234 86 - 11422946 ------------UGGAUGAGGUUUAGAUGAAUGAUAGAAAACA-----UUGAACAAAGUAAAAUAUUUGUUCAAGGUGUUUGAACUGCCAAC-----UAAAUGGAAUC----- ------------(((....(((((((((............((.-----(((((((((........))))))))).))))))))))).)))..-----...........----- ( -17.60, z-score = -2.34, R) >consensus __GGU_UUUGGUUGGGUACGGGCCAAAUACAAGA_AAAAAAAA____GCUGAGCAAAGUAAAAUAUUUGUUCAAAGUGUUUGAACUCGAAGCGCAGGCAAACGAAAUC_____ .................................................((((((((........))))))))...((((((...............)))))).......... (-10.13 = -9.38 + -0.75)

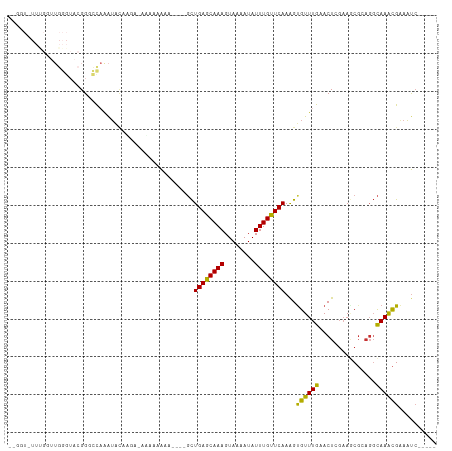
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:12 2011