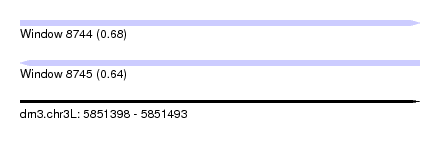
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,851,398 – 5,851,493 |
| Length | 95 |
| Max. P | 0.684547 |

| Location | 5,851,398 – 5,851,493 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Shannon entropy | 0.33934 |
| G+C content | 0.48835 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
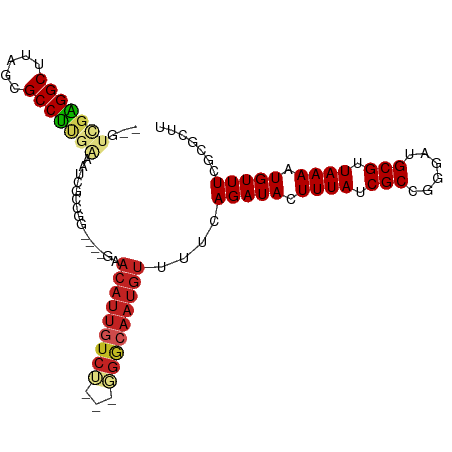
>dm3.chr3L 5851398 95 + 24543557 AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCCC--AGACAAUGUUC----CCGGCGAUUUCAAGGCGCUAAGCCUCGAC-- .(((((.....((((((.(((......))).))))))...((((...((((((..--.(((...))).----..))))))))))..)))))..........-- ( -22.40, z-score = -0.72, R) >droSim1.chr3L 5344510 95 + 22553184 AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCCC--AGACAAUGUUC----CCGGCGAUUUCAAGGCGCUAAGCCUCGAC-- .(((((.....((((((.(((......))).))))))...((((...((((((..--.(((...))).----..))))))))))..)))))..........-- ( -22.40, z-score = -0.72, R) >droSec1.super_2 5787677 95 + 7591821 AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCCC--AGACAAUGUUC----CCGGCGAUUUCAAGGCGCUAAGCCUCGAC-- .(((((.....((((((.(((......))).))))))...((((...((((((..--.(((...))).----..))))))))))..)))))..........-- ( -22.40, z-score = -0.72, R) >droYak2.chr3L 6426004 95 + 24197627 AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCCC--AGACAAUGUUC----CCGGCGAUUUCAAGGCGCUAAGCCUCGAC-- .(((((.....((((((.(((......))).))))))...((((...((((((..--.(((...))).----..))))))))))..)))))..........-- ( -22.40, z-score = -0.72, R) >droEre2.scaffold_4784 8544720 95 + 25762168 AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCCC--AGACAAUGUUC----CCGGCGAUUUCAAGGCGCUAAGCCUCGAC-- .(((((.....((((((.(((......))).))))))...((((...((((((..--.(((...))).----..))))))))))..)))))..........-- ( -22.40, z-score = -0.72, R) >droAna3.scaffold_13337 10607723 83 - 23293914 AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCC------------------CAGACAAUUCCAAGGCGCUAAGCCUCAAC-- .(((((.....((((((.(((......))).)))))).((((............------------------))))..........)))))..........-- ( -16.90, z-score = -1.35, R) >dp4.chrXR_group8 5691371 98 - 9212921 AAGCGCGAAACAUUUUAACGCAUCUCAGCGAUAAAGUAUCUGAAAACAUUGCCC---AGACAAUGUCCAGAACAGCCCAUUCCCAGGCGCUAAGCCUCAAC-- .(((((.....((((((.(((......))).)))))).((((...((((((...---...)))))).))))...............)))))..........-- ( -23.20, z-score = -2.99, R) >droPer1.super_65 73395 98 - 397436 AAGCGCGAAACAUUUUAACGCAUCUCAGCGAUAAAGUAUCUGAAAACAUUGCCC---AGACAAUGUCCAGAACAGCCCAUUCCCAGGCGCUAAGCCUCAAC-- .(((((.....((((((.(((......))).)))))).((((...((((((...---...)))))).))))...............)))))..........-- ( -23.20, z-score = -2.99, R) >droMoj3.scaffold_6680 11056401 92 + 24764193 AAGCGCGAAACAUUUUAACGCGUCGCUGCGAUAAAGUAUCUGAAAACAUUUUCGAACCGACAAUGU---------CCAAUU--GGGGCGCUAAGCCCCUGGAC ..(((..((.....))..)))((((...(((.((.((........)).)).)))...))))...((---------(((...--(((((.....)))))))))) ( -27.00, z-score = -1.99, R) >droVir3.scaffold_13049 15514626 89 - 25233164 AAGCGCGAAACAUUUUAACGCGUCGCGGCGAUAAAGUAUCUGAAAACAUUGCC---CCGACAAUGU---------CCAAUU--GGGGCGCUAAGCCCCUCGAC ..(((..((.....))..)))((((.((((((...((........))))))))---.))))...((---------(.....--(((((.....)))))..))) ( -26.40, z-score = -1.43, R) >droGri2.scaffold_15110 4281243 89 - 24565398 AAGCGCGAAACAUUUUAACGCGUCGAGGCGAUAAAGUAUCUGAAAACAUUGCC---CCGACAAUGU---------CCAAUU--GAGGCGCUAAGCCCCUCGAC ..(((..((.....))..)))((((.((((((...((........))))))))---.)))).....---------....((--((((.((...)).)))))). ( -23.10, z-score = -1.57, R) >consensus AAGCGCGAAACAUUUUAACGCAUCCCGGCGAUAAAGUAUCUGAAAACAUUGCCC___AGACAAUGUUC____CCGCCGAUUUCAAGGCGCUAAGCCUCGAC__ .(((((.....((((((.(((......))).))))))........((((((.(.....).))))))....................)))))............ (-16.42 = -17.15 + 0.73)

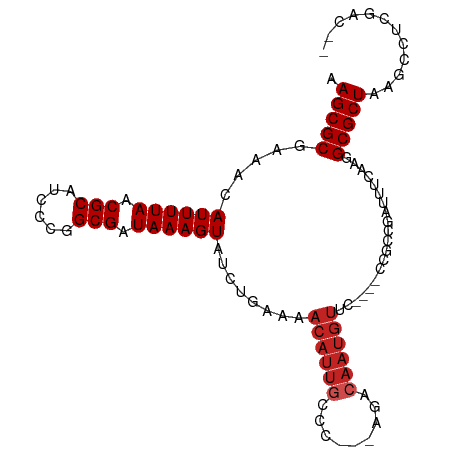
| Location | 5,851,398 – 5,851,493 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.23 |
| Shannon entropy | 0.33934 |
| G+C content | 0.48835 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5851398 95 - 24543557 --GUCGAGGCUUAGCGCCUUGAAAUCGCCGG----GAACAUUGUCU--GGGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU --.(((((((.....))))))).(((.((((----(((((((((..--...)))))))))...((((...)))).)))))))((((.((.....)).)))).. ( -32.70, z-score = -1.80, R) >droSim1.chr3L 5344510 95 - 22553184 --GUCGAGGCUUAGCGCCUUGAAAUCGCCGG----GAACAUUGUCU--GGGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU --.(((((((.....))))))).(((.((((----(((((((((..--...)))))))))...((((...)))).)))))))((((.((.....)).)))).. ( -32.70, z-score = -1.80, R) >droSec1.super_2 5787677 95 - 7591821 --GUCGAGGCUUAGCGCCUUGAAAUCGCCGG----GAACAUUGUCU--GGGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU --.(((((((.....))))))).(((.((((----(((((((((..--...)))))))))...((((...)))).)))))))((((.((.....)).)))).. ( -32.70, z-score = -1.80, R) >droYak2.chr3L 6426004 95 - 24197627 --GUCGAGGCUUAGCGCCUUGAAAUCGCCGG----GAACAUUGUCU--GGGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU --.(((((((.....))))))).(((.((((----(((((((((..--...)))))))))...((((...)))).)))))))((((.((.....)).)))).. ( -32.70, z-score = -1.80, R) >droEre2.scaffold_4784 8544720 95 - 25762168 --GUCGAGGCUUAGCGCCUUGAAAUCGCCGG----GAACAUUGUCU--GGGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU --.(((((((.....))))))).(((.((((----(((((((((..--...)))))))))...((((...)))).)))))))((((.((.....)).)))).. ( -32.70, z-score = -1.80, R) >droAna3.scaffold_13337 10607723 83 + 23293914 --GUUGAGGCUUAGCGCCUUGGAAUUGUCUG------------------GGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU --((.(((((((((((((((((...((((((------------------..(......)..))))))........)))))..)))))))...))))))).... ( -22.30, z-score = -0.23, R) >dp4.chrXR_group8 5691371 98 + 9212921 --GUUGAGGCUUAGCGCCUGGGAAUGGGCUGUUCUGGACAUUGUCU---GGGCAAUGUUUUCAGAUACUUUAUCGCUGAGAUGCGUUAAAAUGUUUCGCGCUU --.......((((((((((......)))).((((((((((((((..---..))))))))..)))).))......))))))..((((.((.....)).)))).. ( -29.70, z-score = -1.20, R) >droPer1.super_65 73395 98 + 397436 --GUUGAGGCUUAGCGCCUGGGAAUGGGCUGUUCUGGACAUUGUCU---GGGCAAUGUUUUCAGAUACUUUAUCGCUGAGAUGCGUUAAAAUGUUUCGCGCUU --.......((((((((((......)))).((((((((((((((..---..))))))))..)))).))......))))))..((((.((.....)).)))).. ( -29.70, z-score = -1.20, R) >droMoj3.scaffold_6680 11056401 92 - 24764193 GUCCAGGGGCUUAGCGCCCC--AAUUGG---------ACAUUGUCGGUUCGAAAAUGUUUUCAGAUACUUUAUCGCAGCGACGCGUUAAAAUGUUUCGCGCUU ((((((((((.....)))))--...)))---------))...((((.(.(((.((.((........)).)).))).).))))((((.((.....)).)))).. ( -29.70, z-score = -2.10, R) >droVir3.scaffold_13049 15514626 89 + 25233164 GUCGAGGGGCUUAGCGCCCC--AAUUGG---------ACAUUGUCGG---GGCAAUGUUUUCAGAUACUUUAUCGCCGCGACGCGUUAAAAUGUUUCGCGCUU (((..(((((.....)))))--....((---------(((((((...---.)))))))))...)))........((.((((.((((....)))).)))))).. ( -30.90, z-score = -1.58, R) >droGri2.scaffold_15110 4281243 89 + 24565398 GUCGAGGGGCUUAGCGCCUC--AAUUGG---------ACAUUGUCGG---GGCAAUGUUUUCAGAUACUUUAUCGCCUCGACGCGUUAAAAUGUUUCGCGCUU ((((((((((.....)))((--....((---------(((((((...---.)))))))))...))..........)))))))((((.((.....)).)))).. ( -29.50, z-score = -2.00, R) >consensus __GUCGAGGCUUAGCGCCUUGAAAUCGCCGG____GAACAUUGUCU___GGGCAAUGUUUUCAGAUACUUUAUCGCCGGGAUGCGUUAAAAUGUUUCGCGCUU ...(((((((.....)))))))...............((((((((.....))))))))....(((((.((((.(((......))).)))).)))))....... (-20.32 = -21.05 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:10 2011