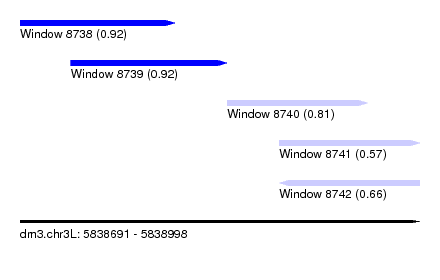
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,838,691 – 5,838,998 |
| Length | 307 |
| Max. P | 0.921321 |

| Location | 5,838,691 – 5,838,810 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.06 |
| Shannon entropy | 0.07159 |
| G+C content | 0.47986 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -33.40 |
| Energy contribution | -35.88 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.921321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5838691 119 + 24543557 UUUCGUAUAAUUCUUUUUUU-AUUUGGACGAACUCGCUCUACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAG .(((((.(((..........-..))).))))).((((((....)))))).((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).. ( -38.40, z-score = -2.67, R) >droSim1.chr3L 5331341 119 + 22553184 UUUCGUAUAAUUCUUUUUUU-AUUUGGGCGAAAUCGCUCUACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAG ((((((.(((..........-..))).))))))((((((....)))))).((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).. ( -39.70, z-score = -2.82, R) >droSec1.super_2 5775122 119 + 7591821 UUUCGUAUAAUUCUUUUUUU-AUUUGGGCGAACUCGCUCUACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAG .(((((.(((..........-..))).))))).((((((....)))))).((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).. ( -38.70, z-score = -2.46, R) >droYak2.chr3L 6412205 120 + 24197627 UUUCGUAUAAUUCUUUUUUUUAUUUGGGCGAACUCGCUCUGCGGAGCGGCACAGGUCUGCUCGAUUUGAUGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCACUGUGGUAG .(((((.(((.............))).)))))...((((((((..((((((((((((.....)))))).))))))((..(((.......)))...)).....)))))))).......... ( -36.02, z-score = -1.35, R) >droEre2.scaffold_4784 8531624 119 + 25762168 UUUCGUAUAAUUCUUUUUUU-AUUUGGGCGAACUCGCUCUACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACAGAUGCAGAGCAGCGCUGUGG .(((((.(((..........-..))).))))).((((((....))))))((((((.(((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).))))). ( -39.90, z-score = -2.40, R) >consensus UUUCGUAUAAUUCUUUUUUU_AUUUGGGCGAACUCGCUCUACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAG .(((((.(((.............))).))))).((((((....)))))).((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).. (-33.40 = -35.88 + 2.48)

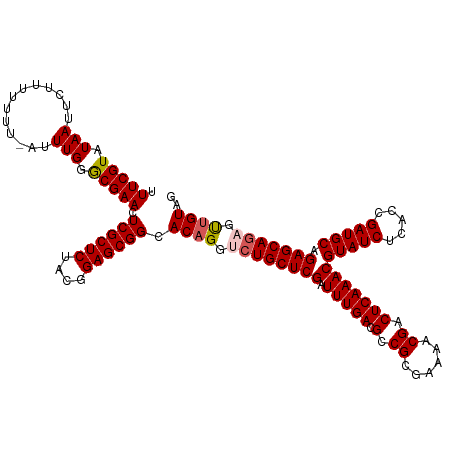
| Location | 5,838,730 – 5,838,850 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Shannon entropy | 0.06557 |
| G+C content | 0.50667 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -33.68 |
| Energy contribution | -34.32 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.921041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5838730 120 + 24543557 ACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAGCAUUCACAACGUAUCUCACGGAUACAACUCAAACGUAUCU ..((.(((((((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).)).........(((((.....)))))........))).)). ( -36.60, z-score = -2.19, R) >droSim1.chr3L 5331380 120 + 22553184 ACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAGCAUUCACAACGUAUCUCACGGAUACAACUCAAACGUAUCU ..((.(((((((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).)).........(((((.....)))))........))).)). ( -36.60, z-score = -2.19, R) >droSec1.super_2 5775161 120 + 7591821 ACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAGCAUUCACAACGUAUCUCACGGAUACAACUCAUACGUAUCU ..((.(((((((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).)).........(((((.....)))))........))).)). ( -36.60, z-score = -2.11, R) >droYak2.chr3L 6412245 120 + 24197627 GCGGAGCGGCACAGGUCUGCUCGAUUUGAUGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCACUGUGGUAGCAUUCACAACGUAUCUCACGGAUACAACUCAAACGUAUCU ..(((((((((((((((.....)))))).))))))....(((........(((((.....))))).(((...(((((......)))))..(((((.....)))))..)))...))).))) ( -36.80, z-score = -1.95, R) >droEre2.scaffold_4784 8531663 120 + 25762168 ACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACAGAUGCAGAGCAGCGCUGUGGCAUUCCCAACGUAUCUCACGGAUACAACUCAAACGUAUCU ..((((.(.((((((.(((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).))))).).))))....(((((.....)))))............... ( -38.40, z-score = -2.14, R) >consensus ACGGAGCGGCACAGGUCUGCUCGAUUUGACGCCGCGAAAACGACUCAAACGUAUCUCACCGAUGCAGAGCAGAGUUGUAGCAUUCACAACGUAUCUCACGGAUACAACUCAAACGUAUCU ..((.(((((((((.((((((((.(((((.(.((......)).)))))))(((((.....))))).))))))).)))).)).........(((((.....)))))........))).)). (-33.68 = -34.32 + 0.64)

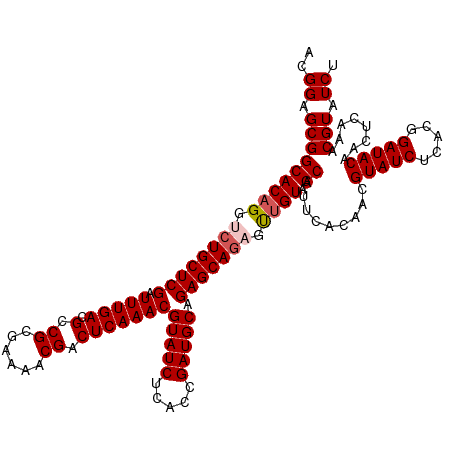
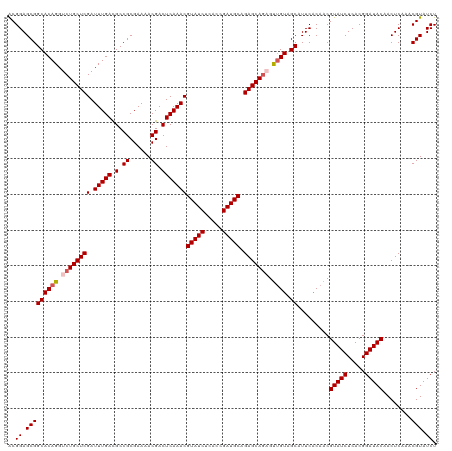
| Location | 5,838,850 – 5,838,958 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.02 |
| Shannon entropy | 0.13256 |
| G+C content | 0.47222 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -27.28 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5838850 108 + 24543557 CACGACUCAAACGUAUCUCACUGGUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCAUUUCGCUCAAACGUUGAGUGUUUUCUCCAAC------------ ....((((((.(((........(((((((.....)))))))(((...(..(((((.((((....)))))))))..)....)))....)))))))))............------------ ( -29.00, z-score = -1.24, R) >droSim1.chr3L 5331500 108 + 22553184 CACGACUCAAACGUAUCUCACUGGUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCUCCAAC------------ ....((((((.(((........(((((((.....)))))))(((...(..(((((.((((....)))))))))..)....)))....)))))))))............------------ ( -29.00, z-score = -1.29, R) >droSec1.super_2 5775281 108 + 7591821 CACGACUCAAACGUAUCUCACUGGUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCUCCAAC------------ ....((((((.(((........(((((((.....)))))))(((...(..(((((.((((....)))))))))..)....)))....)))))))))............------------ ( -29.00, z-score = -1.29, R) >droYak2.chr3L 6412365 120 + 24197627 CACAACUCAAACGUAUCUCACUGGUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACACUCAUUUCGCUCAAACGUUGAGUGUUUUUUUUUUUUUUUGGUUUAAC ...(((.((((............((((((...(((((((..(((......)))..)))))))..))))))(((((((...((......)).)))))))...........))))))).... ( -28.10, z-score = -1.46, R) >droEre2.scaffold_4784 8531783 108 + 25762168 CACGACUCAAACGUAUCUCACUGGUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACACUCUUUUCGCUCAAACGUUGAGUGUUUUCUUCAAC------------ ....((((((.(((........(((((((.....)))))))(((...((..(((((((((....))))).)))).))...)))....)))))))))............------------ ( -27.00, z-score = -1.08, R) >consensus CACGACUCAAACGUAUCUCACUGGUCGGAAUUUGUUCGACUGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCUCCAAC____________ ....((((((.(((........(((((((.....)))))))(((...((.(((((.((((....))))))))).))....)))....)))))))))........................ (-27.28 = -27.68 + 0.40)

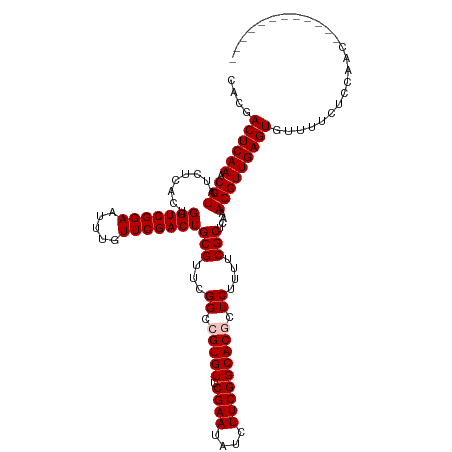
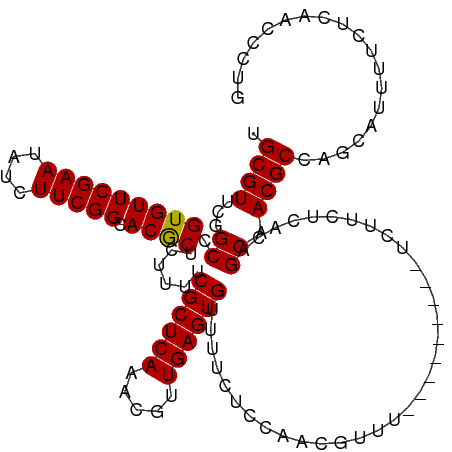
| Location | 5,838,890 – 5,838,998 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.86 |
| Shannon entropy | 0.15447 |
| G+C content | 0.47510 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.36 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5838890 108 + 24543557 UGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCAUUUCGCUCAAACGUUGAGUGUUUUCUCCAACGUUU---------UCUUCUCACAGCAACGCCAGCAUUUUCUCAACCCUG ...(((.(((.((((.((((....))))))))(((..........((((((((((......)).))))))))---------.........)))...))))))............... ( -24.31, z-score = -1.70, R) >droSim1.chr3L 5331540 108 + 22553184 UGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCUCCAACGUUU---------UCUUCUCGCAGCAACGCCAGCAUUUUCUCAACCCUG ...(((.(((.((((.((((....))))))))(((..........((((((((((......)).))))))))---------.........)))...))))))............... ( -24.31, z-score = -1.16, R) >droSec1.super_2 5775321 108 + 7591821 UGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCUCCAACGUUU---------UCUUCUCACAGCAACGCCAGCAUUUUCUCAACCCUG ...(((.(((.((((.((((....))))))))(((..........((((((((((......)).))))))))---------.........)))...))))))............... ( -24.31, z-score = -1.71, R) >droYak2.chr3L 6412405 117 + 24197627 UGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACACUCAUUUCGCUCAAACGUUGAGUGUUUUUUUUUUUUUUUGGUUUAACGUUUUCUCACAGCAACGCCAGCAUUUUCUCAACCCUG .(((((..((((((((((((....))))).))))......((((((.....))))))...............)))..))))).........((.......))............... ( -24.20, z-score = -1.36, R) >droEre2.scaffold_4784 8531823 105 + 25762168 UGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACACUCUUUUCGCUCAAACGUUGAGUGUUUUCUUCAACGUUU---------UC---UCACAGCAACGCCAUCAUUUUCUCAACCCUG .......(((.(((((((((....))))).)))).......(((.((((((((((.......))))))))))---------..---....)))...))).................. ( -23.00, z-score = -2.34, R) >consensus UGCGUUCGGCCGUGUUCGAAUAUCUUCGGCACGCUCUUUUCGCUCAAACGUUGAGUGUUUUCUCCAACGUUU_________UCUUCUCACAGCAACGCCAGCAUUUUCUCAACCCUG .((((...((.(((((((((....))))).))))......((((((.....))))))..................................)).))))................... (-19.60 = -19.36 + -0.24)

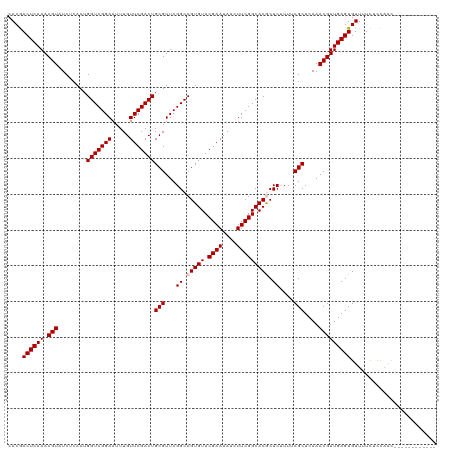
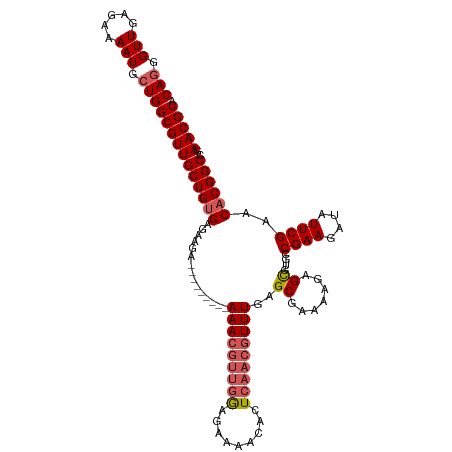
| Location | 5,838,890 – 5,838,998 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.86 |
| Shannon entropy | 0.15447 |
| G+C content | 0.47510 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.54 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5838890 108 - 24543557 CAGGGUUGAGAAAAUGCUGGCGUUGCUGUGAGAAGA---------AAACGUUGGAGAAAACACUCAACGUUUGAGCGAAAUGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCA (((.(((.....))).)))(((((((((((......---------((((((((.((......))))))))))..((.......))....((((....))))..))))))..))))). ( -32.40, z-score = -1.86, R) >droSim1.chr3L 5331540 108 - 22553184 CAGGGUUGAGAAAAUGCUGGCGUUGCUGCGAGAAGA---------AAACGUUGGAGAAAACACUCAACGUUUGAGCGAAAAGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCA (((.(((.....))).)))(((((.(.((.....(.---------((((((((.((......))))))))))...)........((((.((((....))))..)))))).)))))). ( -29.70, z-score = -0.72, R) >droSec1.super_2 5775321 108 - 7591821 CAGGGUUGAGAAAAUGCUGGCGUUGCUGUGAGAAGA---------AAACGUUGGAGAAAACACUCAACGUUUGAGCGAAAAGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCA (((.(((.....))).)))(((((((((((......---------((((((((.((......))))))))))..((.......))....((((....))))..))))))..))))). ( -32.40, z-score = -1.88, R) >droYak2.chr3L 6412405 117 - 24197627 CAGGGUUGAGAAAAUGCUGGCGUUGCUGUGAGAAAACGUUAAACCAAAAAAAAAAAAAAACACUCAACGUUUGAGCGAAAUGAGUGUGCCGAAGAUAUUCGAACACGGCCGAACGCA (((.(((.....))).)))(((((((((((.............................(((((((.(((....)))...)))))))..((((....))))..))))))..))))). ( -32.80, z-score = -2.75, R) >droEre2.scaffold_4784 8531823 105 - 25762168 CAGGGUUGAGAAAAUGAUGGCGUUGCUGUGA---GA---------AAACGUUGAAGAAAACACUCAACGUUUGAGCGAAAAGAGUGUGCCGAAGAUAUUCGAACACGGCCGAACGCA ((..(((.....)))..))(((((((((((.---(.---------(((((((((.........)))))))))...).....((((((.......))))))...))))))..))))). ( -28.40, z-score = -1.58, R) >consensus CAGGGUUGAGAAAAUGCUGGCGUUGCUGUGAGAAGA_________AAACGUUGGAGAAAACACUCAACGUUUGAGCGAAAAGAGCGUGCCGAAGAUAUUCGAACACGGCCGAACGCA (((.(((.....))).)))(((((((((((...............(((((((((.........)))))))))..((.......))....((((....))))..))))))..))))). (-25.62 = -26.54 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:07 2011