
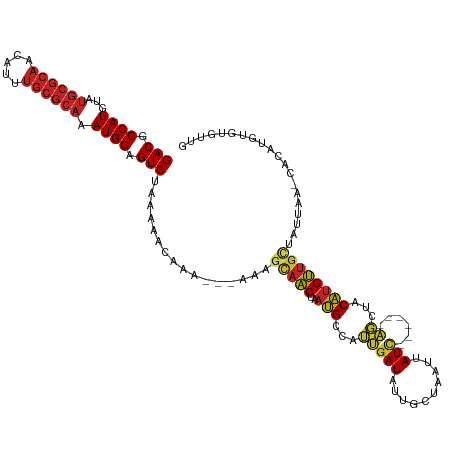
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,836,550 – 5,836,664 |
| Length | 114 |
| Max. P | 0.940758 |

| Location | 5,836,550 – 5,836,664 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.49181 |
| G+C content | 0.40936 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.780878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
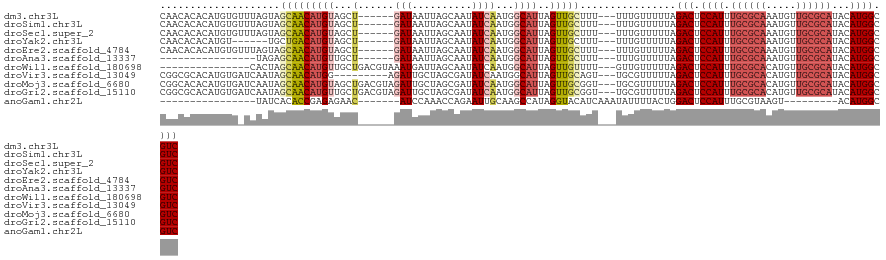
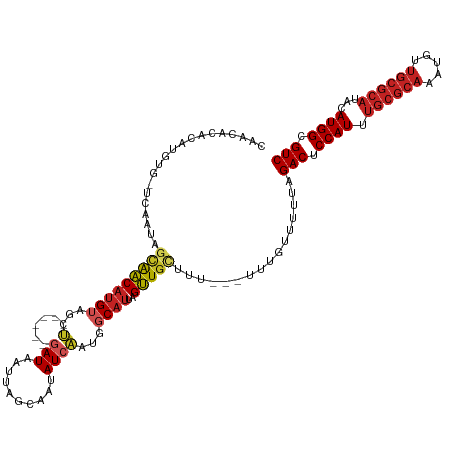
>dm3.chr3L 5836550 114 + 24543557 CAACACACAUGUGUUUAGUAGCAACAUGUAGCU------GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU---UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC .(((((....)))))....((((((((((...(------((((........)))))...))))..))))))..---..........(((.((((.((((((.....))))))...)))).))) ( -31.70, z-score = -1.64, R) >droSim1.chr3L 5329226 114 + 22553184 CAACACACAUGUGUUUAGUAGCAACAUGUAGCU------GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU---UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC .(((((....)))))....((((((((((...(------((((........)))))...))))..))))))..---..........(((.((((.((((((.....))))))...)))).))) ( -31.70, z-score = -1.64, R) >droSec1.super_2 5773003 114 + 7591821 CAACACACAUGUGUUUAGUAGCAACAUGUAGCU------GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU---UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC .(((((....)))))....((((((((((...(------((((........)))))...))))..))))))..---..........(((.((((.((((((.....))))))...)))).))) ( -31.70, z-score = -1.64, R) >droYak2.chr3L 6409935 108 + 24197627 CAACACACAUGU------UGCUGACAUGUAGCU------GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU---UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC ......((((((------(...))))))).(((------((...))))).....(((.((((.....))))..---.)))......(((.((((.((((((.....))))))...)))).))) ( -28.90, z-score = -1.05, R) >droEre2.scaffold_4784 8529461 114 + 25762168 CAACACACAUGUGUUUAGUAGCAACAUGUAGCU------GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU---UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC .(((((....)))))....((((((((((...(------((((........)))))...))))..))))))..---..........(((.((((.((((((.....))))))...)))).))) ( -31.70, z-score = -1.64, R) >droAna3.scaffold_13337 10590892 98 - 23293914 ----------------UAGAGCAACAUGUUGCU------GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU---UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC ----------------.(((((((((((((..(------((((........)))))..)))))..))))))))---..........(((.((((.((((((.....))))))...)))).))) ( -31.80, z-score = -3.10, R) >droWil1.scaffold_180698 9338216 105 + 11422946 ---------------CACUAGCAACAUGUUGCUGACGUAAAUGAUUAGCAAUAUCAAUGGCAUUAGUUGUUUU---GUUGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUC ---------------....((((((((((((((((((....)).)))))))))..((..((....))..)).)---))))))....(((.((((.((((((.....))))))...)))).))) ( -31.60, z-score = -1.90, R) >droVir3.scaffold_13049 15495941 111 - 25233164 CGGCGCACAUGUGAUCAAUAGCAACAUGG---------AGAUUGCUAGCGAUAUCAAUGGCAUUAGUUGCAGU---UGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUC .(((((.((((((............((((---------((.(((..((((....((((.(((.....))).))---))))))..))).)))))).((((((.....))))))))))))))))) ( -38.40, z-score = -1.90, R) >droMoj3.scaffold_6680 11037741 120 + 24764193 CGGCACACAUGUGAUCAAUAGCAACAUGUAGCUGACGUAGAUUGCUAGCGAUAUCAAUGGCAUUAGUUGCGGU---UGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUC .(((((....))).))....(((((.(((((((((.((..((((.((....)).)))).)).)))))))))))---))).......(((.((((.((((((.....))))))...)))).))) ( -38.70, z-score = -0.98, R) >droGri2.scaffold_15110 4262105 120 - 24565398 CGGCGCACAUGUGAUCAAUAGCAACAUGUUGCUGACGUAGAUUGCUAGCGAUAUCAAUGGCAUUAGUUGCGGU---UGCGUUUUUAGACUCCAUUUGCGCACAUGUUGCGCAUACAUGGCGUC .(((((.((((((.......((((((((((((((.((.....)).))))))))).....(((.....))).))---)))................((((((.....))))))))))))))))) ( -41.10, z-score = -1.11, R) >anoGam1.chr2L 6449847 91 - 48795086 ----------------UAUCACACCGAGAGAAC-------AUCCAAACCAGAAUUGCAAGCCAUAGGUACAUCAAAUAUUUUACUGGACUCCAUUUGCGUAAGU---------ACAUGGCGUC ----------------.................-------.......((((........(((...)))......((....)).))))((.((((.(((....))---------).)))).)). ( -12.70, z-score = 0.53, R) >consensus CAACACACAUGUG_UCAAUAGCAACAUGUAGCU______GAUAAUUAGCAAUAUCAAUGGCAUUAGUUGCUUU___UUUGUUUUUAGACUCCAUUUGCGCAAAUGUUGCGCAUACAUGGCGUC ....................(((((((((..............................))))..)))))................(((.((((.((((((.....))))))...)))).))) (-17.08 = -17.87 + 0.80)
| Location | 5,836,550 – 5,836,664 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.49181 |
| G+C content | 0.40936 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.51 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.940758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
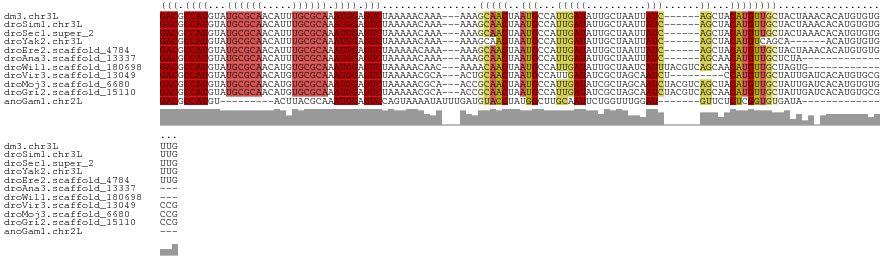
>dm3.chr3L 5836550 114 - 24543557 GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA---AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC------AGCUACAUGUUGCUACUAAACACAUGUGUGUUG (((.((((...((((((.....)))))).)))).)))..........---..((((((....((...(((((........))))------))).....))))))....(((((....))))). ( -33.40, z-score = -3.14, R) >droSim1.chr3L 5329226 114 - 22553184 GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA---AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC------AGCUACAUGUUGCUACUAAACACAUGUGUGUUG (((.((((...((((((.....)))))).)))).)))..........---..((((((....((...(((((........))))------))).....))))))....(((((....))))). ( -33.40, z-score = -3.14, R) >droSec1.super_2 5773003 114 - 7591821 GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA---AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC------AGCUACAUGUUGCUACUAAACACAUGUGUGUUG (((.((((...((((((.....)))))).)))).)))..........---..((((((....((...(((((........))))------))).....))))))....(((((....))))). ( -33.40, z-score = -3.14, R) >droYak2.chr3L 6409935 108 - 24197627 GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA---AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC------AGCUACAUGUCAGCA------ACAUGUGUGUUG (((.((((...((((((.....)))))).)))).)))....((((..---...(((.....)))..((((((........))))------)).(((((((.....------))))))))))). ( -28.70, z-score = -1.57, R) >droEre2.scaffold_4784 8529461 114 - 25762168 GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA---AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC------AGCUACAUGUUGCUACUAAACACAUGUGUGUUG (((.((((...((((((.....)))))).)))).)))..........---..((((((....((...(((((........))))------))).....))))))....(((((....))))). ( -33.40, z-score = -3.14, R) >droAna3.scaffold_13337 10590892 98 + 23293914 GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA---AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC------AGCAACAUGUUGCUCUA---------------- (((.((((...((((((.....)))))).)))).)))..........---..((((((...(((...(((((........))))------))))....))))))...---------------- ( -29.60, z-score = -3.73, R) >droWil1.scaffold_180698 9338216 105 - 11422946 GACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACAAC---AAAACAACUAAUGCCAUUGAUAUUGCUAAUCAUUUACGUCAGCAACAUGUUGCUAGUG--------------- (((.((((...((((((.....)))))).)))).)))..........---.................((((.......))))....((..(((((....)))))..))--------------- ( -24.00, z-score = -0.41, R) >droVir3.scaffold_13049 15495941 111 + 25233164 GACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCA---ACUGCAACUAAUGCCAUUGAUAUCGCUAGCAAUCU---------CCAUGUUGCUAUUGAUCACAUGUGCGCCG (((.((((...((((((.....)))))).)))).)))......((((---...(((.....)))((((((..(((.(((((((..---------....))))))).)))))).)))))))... ( -33.00, z-score = -1.55, R) >droMoj3.scaffold_6680 11037741 120 - 24764193 GACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCA---ACCGCAACUAAUGCCAUUGAUAUCGCUAGCAAUCUACGUCAGCUACAUGUUGCUAUUGAUCACAUGUGUGCCG (((.((((...((((((.....)))))).)))).))).....(((((---...(((.....)))...((..((((.(((((((...............))))))).))))..))))))).... ( -30.96, z-score = 0.15, R) >droGri2.scaffold_15110 4262105 120 + 24565398 GACGCCAUGUAUGCGCAACAUGUGCGCAAAUGGAGUCUAAAAACGCA---ACCGCAACUAAUGCCAUUGAUAUCGCUAGCAAUCUACGUCAGCAACAUGUUGCUAUUGAUCACAUGUGCGCCG (((.((((...((((((.....)))))).)))).)))......((((---...(((.....)))((((((..(((.(((((((....((.....))..))))))).)))))).)))))))... ( -31.80, z-score = 0.05, R) >anoGam1.chr2L 6449847 91 + 48795086 GACGCCAUGU---------ACUUACGCAAAUGGAGUCCAGUAAAAUAUUUGAUGUACCUAUGGCUUGCAAUUCUGGUUUGGAU-------GUUCUCUCGGUGUGAUA---------------- .(((((.(((---------......)))...((((..((...((((......((((((...))..))))......))))...)-------)..)))).)))))....---------------- ( -15.60, z-score = 1.48, R) >consensus GACGCCAUGUAUGCGCAACAUUUGCGCAAAUGGAGUCUAAAAACAAA___AAAGCAACUAAUGCCAUUGAUAUUGCUAAUUAUC______AGCUACAUGUUGCUAUUAA_CACAUGUGUGUUG (((.((((...((((((.....)))))).)))).)))................(((((........................................))))).................... (-17.90 = -18.51 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:03 2011