| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,408,424 – 4,408,534 |
| Length | 110 |
| Max. P | 0.510071 |

| Location | 4,408,424 – 4,408,534 |
|---|---|
| Length | 110 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.53 |
| Shannon entropy | 0.64904 |
| G+C content | 0.54597 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -17.27 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.510071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
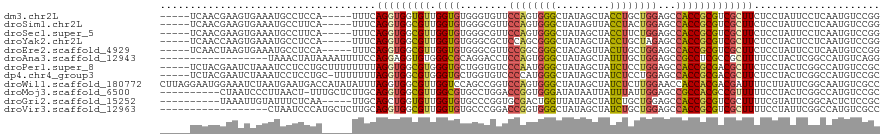
>dm3.chr2L 4408424 110 - 23011544 -----UCAACGAAGUGAAAUGCCUCCA-----UUUCAGGUGGUGUUGGUGUGGGUGUUCCAGUGGGCUAUAGCUACCUGCUGGAGCCACCGCGUCGCUUCUCCUAUUCCUCAAUGUCCGG -----.....(((((((((((((.((.-----.....)).))))))...((((..(((((((..((.........))..)))))))..)))).))))))).................... ( -41.00, z-score = -2.51, R) >droSim1.chr2L 4332848 110 - 22036055 -----UCAACGAAGUGAAAUGCCUUCA-----UUUCAGGUGGCGUUGGUGUGGGCGUUCCAGUGGGCUAUAGUUACCUACUGGAGCCACCGCGUCGCUUCUCCUAUUCCUCAAUGUCCGG -----.....(((((((((((((.((.-----.....)).))))))...((((..(((((((((((.((....)))))))))))))..)))).))))))).................... ( -41.50, z-score = -2.94, R) >droSec1.super_5 2506710 110 - 5866729 -----UCAACGAAGUGAAAUGCCUUCA-----UUUCAGGUGGCGUUGGUGUGGGCGUUCCAGUGGGCUAUAGCUACCUUCUGGAGCCACCGCGUCGCUUCUCCUAUUCCUCAAUGUCCGG -----.....(((((((((((((.((.-----.....)).))))))...((((..(((((((.(((.........))).)))))))..)))).))))))).................... ( -36.70, z-score = -1.39, R) >droYak2.chr2L 4441927 110 - 22324452 -----UCAACCAAGUGAAAUGCCUCCA-----UUUCAGGUGGCGUUGGUGUGGGCGCUCCAGCGGGCUAUAGCUACCUGCUAGAGCCACCGCGUCGCUUCUCCUACUCCUCAAUGUCCGG -----......((((((((((((.((.-----.....)).))))))...((((..((((.((((((.........)))))).))))..)))).))))))..................... ( -37.70, z-score = -1.24, R) >droEre2.scaffold_4929 4490904 110 - 26641161 -----UCAACUAAGUGAAAUGCCUCCA-----UUUCAGGUGGCGUUGGUGUGGGCGUUCCGGCGGGCUACAGUUACUUGCUGGAGCCACCGCGUCGCUUCUCCUAUUCCUCAAUGUCCGG -----......((((((((((((.((.-----.....)).))))))...((((..(((((((((((.........)))))))))))..)))).))))))..................... ( -38.20, z-score = -1.31, R) >droAna3.scaffold_12943 1595601 102 + 5039921 ------------------UAAACUAUAAAAUUUUCCAGGAGGUGUGGGCGCAGGACCUCCAGUGGGCUAUAGCUAUUUGCUGGAGCCGCCUCGCCGCUUUUCCUACUCGGCCAUGUCAGG ------------------................((((((((.((((.((.(((..(((((((.(((....)))....)))))))...))))))))).))))))....(((...))).)) ( -33.80, z-score = -0.44, R) >droPer1.super_8 3429588 115 + 3966273 -----UCUACGAAUCUAAAUCCUCCUGCUUUUUUUUAGGUGGCGUGGGUGCUGGUGUCCCAAUGGGCUAUAGCUAUCUCCUGGAGCCACCGCGACGCUUCUCCUACUCGGCCAUGUCCGC -----....(((........((.((((........)))).)).(((((.(..((((((......((((.(((.......))).)))).....)))))).).))))))))........... ( -29.40, z-score = 0.99, R) >dp4.chr4_group3 2345957 114 + 11692001 -----UCUACGAAUCUAAAUCCUCCUGC-UUUUUUUAGGUGGCGUGGGUGCUGGUGUCCCCAUGGGCUAUAGCUAUCUCCUGGAGCCACCGCGACGCUUCUCCUACUCGGCCAUGUCCGC -----....(((........((.((((.-......)))).)).(((((.(..((((((......((((.(((.......))).)))).....)))))).).))))))))........... ( -29.10, z-score = 1.17, R) >droWil1.scaffold_180772 4795521 120 + 8906247 CUUAGGAAUGGAAAUCUAAUGAAUGACCAUAUAUUUAGGUGGCGUUGGUCCAGCCGGUCCAGUGGGCUAUAGCUAUCUCUUGGAACCACCACGACGAUUUUCUUAUUCGGCAAUGUCGCC .....(((((((((((...((((((......))))))(((((((((((.((....)).))))))(((....)))...........))))).....)))))))..))))(((......))) ( -30.00, z-score = 0.30, R) >droMoj3.scaffold_6500 9541205 109 + 32352404 ----------CUAAUCCCUUAACU-UUUGCUCUUGCAGGUGGCGUUGGCGUGCCUGGACCGGUGGGAUAUAAUUAUUUAUUGGAGCCGCCACGCCGUUUUUCCUACUCGGCCAUGUCCGC ----------.((((.((......-(((((....))))).)).))))(((..(.(((.(((((((((.((((....)))).(((((.((...)).))))))))))).)))))).)..))) ( -29.00, z-score = 0.85, R) >droGri2.scaffold_15252 1974864 105 + 17193109 ----------UAAAUUGUAUUUCUCAA-----UUGCAGCUGGUGUUGGUGUGCCCGGUGCGACUGGUUAUAGCUAUCUGCUGGAGCCACCGCGUCGCUUUUCGUAUUCGGCACUCUCCGC ----------..(((((.......)))-----)).((((....))))(.((((((((.((((((((((.((((.....)))).)))))....)))))...))).....))))).)..... ( -30.60, z-score = -0.21, R) >droVir3.scaffold_12963 2424590 102 + 20206255 ------------------CUAAUCCCAUGCUCUUGCAGGUGGCGUUGGUGUGCCCGGACCGGUGGGCUAUAGCUAUCUGCUGGAGCCACCGCGUCGCUUUUCCUAUUCGGCCAUGUCGCC ------------------......((.(((....))))).((((...(((.(((..(((((((((.((.((((.....)))).)))))))).)))(......).....))))))..)))) ( -36.20, z-score = -0.11, R) >consensus _____UCAACGAAGUGAAAUGCCUCCA_____UUUCAGGUGGCGUUGGUGUGGGCGUUCCAGUGGGCUAUAGCUAUCUGCUGGAGCCACCGCGUCGCUUCUCCUAUUCGGCAAUGUCCGC ....................................(((((((((.((((........((((((((.........))))))))...)))))))))))))..................... (-17.27 = -17.20 + -0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:22 2011