| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,767,820 – 5,767,914 |
| Length | 94 |
| Max. P | 0.918602 |

| Location | 5,767,820 – 5,767,914 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 60.15 |
| Shannon entropy | 0.79619 |
| G+C content | 0.51884 |
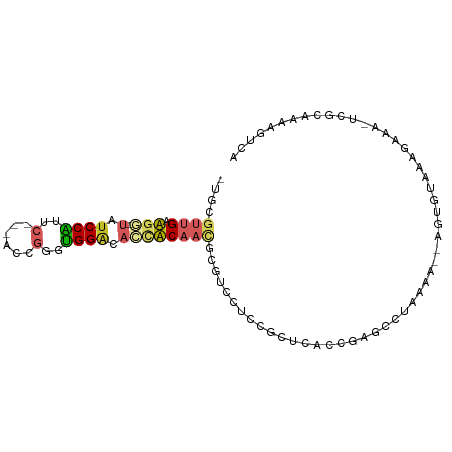
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -7.81 |
| Energy contribution | -7.58 |
| Covariance contribution | -0.23 |
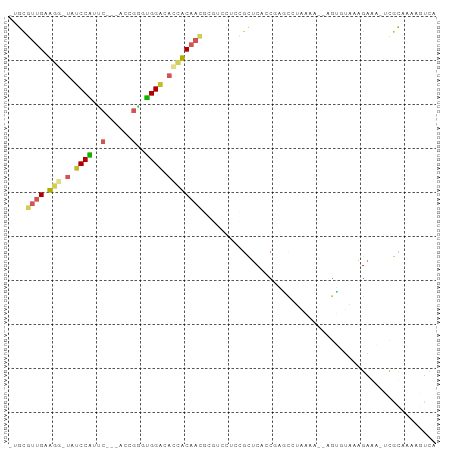
| Combinations/Pair | 1.58 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
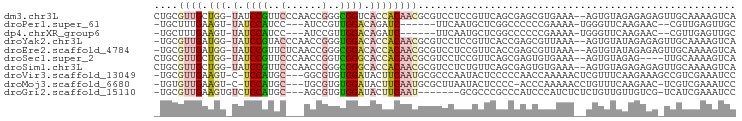
>dm3.chr3L 5767820 94 + 24543557 CUGCGUUGCUGG-UAUCCGUUCCCAACCGGGCGGUCACCACAACGCGUCCUCCGUUCAGCGAGCGUGAAA--AGUGUAGAGAGAGUUGCAAAAGUCA ..((((((.(((-(..((((((......))))))..))))))))))......(((((...)))))(((..--..(((((......)))))....))) ( -30.20, z-score = -1.19, R) >droPer1.super_61 288805 83 - 350175 -UGCUUUGAAGU-UAUCCAUCC---AUCCGUUGGACAGAUC------UUCAAUGCUCGGCCCCCCGAAAA-UGGGUUCAAGAAC--CGUUGAGUUGC -.((.((((((.-.(((..(((---(.....))))..))))------))))).))((((....))))...-..((((....)))--).......... ( -21.00, z-score = -0.78, R) >dp4.chrXR_group6 3528356 83 - 13314419 -UGCUUUGAAGU-UAUCCAUCC---AUCCGUUGGACAGAUC------UUCAAUGCUCGGCCCCCCGAAAA-UGGGUUCAAGAAC--CGUUGAGUUGC -.((.((((((.-.(((..(((---(.....))))..))))------))))).))((((....))))...-..((((....)))--).......... ( -21.00, z-score = -0.78, R) >droYak2.chr3L 6337299 93 + 24197627 -UGCGUUGAUGG-UAUCCGUACCCAACCGGGUGGACACCACAACGCGUCCUCCGUUCACCGAGCGUUAAA--AGUGUAUAGAGAGUUGCAAAAGUCA -.((((((.(((-(.(((..((((....))))))).))))))))))......(((((...))))).....--..((((........))))....... ( -28.80, z-score = -1.82, R) >droEre2.scaffold_4784 8458393 93 + 25762168 -UGCGUUGAUGG-UAUCCGUUCUCAACCGGGCGGACACCACAACGCGUCCUCCGUUCACCGAGCGUUAAA--AGUGUAUAGAGAGUUGCAAAAGUCA -.((((((.(((-(.(((((((......))))))).))))))))))......(((((...))))).....--..((((........))))....... ( -30.30, z-score = -2.14, R) >droSec1.super_2 5704653 90 + 7591821 CUGCGUUGCUGG-UAUCCGUUCCCAACCGGUCGGGCACCACAACGCGUCCUCCGUUCAGCGAGUGUGAAA--AGUGUAGAG----UUGCAAAAGUCA ..((((((.(((-(..(((..((.....)).)))..))))))))))............((((.(.((...--....)).).----))))........ ( -26.60, z-score = -0.75, R) >droSim1.chr3L 5279552 94 + 22553184 CUGCGUUGCUGG-UAUCCGUUCCCAACCGGGCGGGCACCACAACGCGUCCUCUGUUCAGCGAGUGUGAAA--AGUGUAGAGAGAGUUGCAAAAGUCA ..((((((.(((-(..((((((......))))))..))))))))))((.((((.(((.(((..(.....)--..))).)))))))..))........ ( -31.90, z-score = -1.33, R) >droVir3.scaffold_13049 8317792 91 + 25233164 -UGCGUUGAAGU-C-UCCAUGC---GGCGUGUGGAUACUUCAAUGCGCCCAAUACUCCCCCAACCAAAAACUCGUUUCAAGAAAGCCGUCGAAAUCC -.((((((((((-.-(((((((---...))))))).))))))))))...........................(((((............))))).. ( -22.80, z-score = -2.41, R) >droMoj3.scaffold_6680 11971498 89 + 24764193 -UGUGUUGAAGU-C-UCCAUGC---UGCGUGUGGAUACUUCAAUGCGCUUAAUACUCCCC-ACCCAAAAACCUGUUUCAAGAAC-UCGUCGAAAUCC -.((((((((((-.-(((((((---...))))))).))))))))))..............-............(((((..(...-.)...))))).. ( -20.60, z-score = -3.07, R) >droGri2.scaffold_15110 7346586 85 + 24565398 -UGCGUUGAAGUGUCUCCAUGC---AGCGUGUGGAUACUUCAAU-------GCGCCCGCCCAUCCCAUCUCUCUGUUGUUGUCG-UCAUCGAAAUCC -.(((((((((((((..((((.---..))))..)))))))))))-------))............................(((-....)))..... ( -25.30, z-score = -3.36, R) >consensus _UGCGUUGAAGG_UAUCCAUUC___ACCGGGUGGACACCACAACGCGUCCUCCGCUCACCGAGCCUAAAA__AGUGUAAAGAAA_UCGCAAAAGUCA ....((((.(((...((((..(......)..))))..)))))))..................................................... ( -7.81 = -7.58 + -0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:56 2011