
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,765,213 – 5,765,359 |
| Length | 146 |
| Max. P | 0.553438 |

| Location | 5,765,213 – 5,765,332 |
|---|---|
| Length | 119 |
| Sequences | 15 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.80 |
| Shannon entropy | 0.61745 |
| G+C content | 0.51370 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -15.01 |
| Energy contribution | -14.27 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 2.09 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5765213 119 + 24543557 GUAUUUGAUGCGGUUUCGUAACUACAAAGAGCUGGAUCAGUUGGUUAAGGAGACCACGCCGCUGGGACAGAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGCAACC ........((((((((........((((((.(((..(((((.(((...(....)...))))))))..))).))))))((((....))))((((((.....))))))..))))))))... ( -43.50, z-score = -1.20, R) >droSim1.chr3L 5276956 119 + 22553184 GUAUCUAAUGCGGUUCCGCAACUACAAAGAGCUGGAUCAGCUGGUAAAGGAGACCACGCCCCUGGGACAGAUCUUUGGGGGCUAUCUGCGCAGCGAGGUGCGGUGUCUGAGCUGCAACC ........((((((((........((((((.(((..((((..(((...(....)...))).))))..))).)))))).((((...((((((......)))))).))))))))))))... ( -44.30, z-score = -0.92, R) >droSec1.super_2 5702054 119 + 7591821 GUAUCUAAUGCGGUUCCGCAACUACAAAGAGCUGGAUCAGCUGGUGAAGGAGACUACGCCGCUGGGACAGAUCUUUGGGGGCUAUCUGCGCAGCGAGGUGCGGUGCCUGAGCUGCAACC ........((((((((........((((((.(((..(((((.((((.((....)).)))))))))..))).)))))).((((...((((((......)))))).))))))))))))... ( -56.30, z-score = -4.18, R) >droEre2.scaffold_4784 8455818 119 + 25762168 CUAUCUGAUGCGGUUCCGCAACUACAAAGAGCUGGAUCAGCUGGUAAAGGAGACUACGCCGCUGGGACAGAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGCCUGAGCUGCAACC ........((((....))))....((((((.(((..(((((.(((..((....))..))))))))..))).))))))((((((....(.((((((.....)))))))..)))))).... ( -53.60, z-score = -3.38, R) >droYak2.chr3L 6334712 119 + 24197627 CUAUCUGAUGCGGUUCCGCAACUACAAAGAGCUGGAUCAGCUGGUAAAGGAGACUACGCCGCUGGGACAAAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGCAACC ........((((....))))....((((((..((..(((((.(((..((....))..))))))))..))..))))))((((((....(.((((((.....)))))))..)))))).... ( -48.50, z-score = -2.52, R) >droAna3.scaffold_13337 10518052 119 - 23293914 GUAUCUCAUGCGCUUUCGUAACUACAAGGAGCUCGAUCAACUGGUUAAGGAGACGACGCCCCUGGGACAGAUCUUCGGCGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGUAACC (((((((.(((((((((.((((((....((......))...))))))..))))....(((((..(((....)))..)).))).....)))))..)))))))(((.....)))....... ( -37.00, z-score = 0.67, R) >dp4.chrXR_group6 3525642 119 - 13314419 GUACUUGAUGCGUUUCCGCAACUUCAAGGAGCUAGAUCAGCUGGUGAAGGAGACGACCCCACUGAAUCAGAUAUUCGGUGGCUACCUGCGCAGCGAGGUGCGUUGCCUCAGCUGCAGCC ...(((((((((....))))...)))))..(((......((.((((..(....)....(((((((((.....))))))))).)))).))((((((((((.....))))).)))))))). ( -47.30, z-score = -2.35, R) >droPer1.super_61 286073 119 - 350175 GUACUUGAUGCGUUUCCGCAACUUCAAGGAGCUAGAUCAGCUGGUGAAGGAGACGACGCCACUGAAUCAGAUAUUCGGUGGCUACCUGCGCAGCGAGGUGCGUUGCCUCAGCUGCAGCC ...(((((((((....))))...)))))..(((......((.((((..(....)...((((((((((.....)))))))))))))).))((((((((((.....))))).)))))))). ( -51.50, z-score = -3.19, R) >droWil1.scaffold_180698 3794396 119 + 11422946 AUAUUUGAUGCGUUAUCGCAACUAUAAGGAAUUGGAUCAAUUGGUUAAGGAGACAACGCCGCUGAAUCAAAUAUUCGGUGGUUAUCUAAGAAGUGAAGUUCGUUGUUUGAGUUGUAAUC .................((((((((((.(((((..((...((((.((.(....)...((((((((((.....)))))))))))).))))...))..))))).))))...)))))).... ( -27.50, z-score = -1.06, R) >droMoj3.scaffold_6680 11968706 119 + 24764193 GUAUCUGAUGCGCUUUCGCAACUACAAAGAACUUGAUCAGCUGGUGAAGGAGACGACGCCGCUGAGUCAAAUCUUUGGUGGCUAUUUGCGUAGCGAGGUUCGCUGCCUUAGCUGCAAUC ........((((....))))....((((((..(((((((((.((((..(....)..))))))))).)))).))))))(..((((...(.((((((.....))))))).))))..).... ( -46.70, z-score = -3.23, R) >droVir3.scaffold_13049 8314899 119 + 25233164 GUAUCUGAUGCGUUUUCGCAAUUUCAAAGAGCUCGAUCAGCUGGUGAAGGAGACGACGCCAAUUAGUCAAAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGCCUCAGCUGCAAUC ........((((....))))....((((((....(((.((.(((((..(....)..))))).)).)))...))))))((((....))))((((((((((.....))))).))))).... ( -42.70, z-score = -1.20, R) >droGri2.scaffold_15110 7343423 119 + 24565398 GUAUCUGAUGCGUUUUCGCAACUACAAAGAGCUCGAUCAGUUGGUGAAAGAGACGACGCCGUUGAAUCAAAUCUUUGGUGGCUAUCUGCGCAGCGAGGUGCGCUGCCUGAGCUGUAAUC ........((((....)))).......(.((((((((..((((.(....)...))))((((..(((.......)))..)))).)))...((((((.....))))))..))))).).... ( -33.00, z-score = 0.83, R) >anoGam1.chr2L 2226350 119 - 48795086 CUACAUCGGCCGAACGAAGAACAGCAAGGAGCUGGACCAGUACAGCAAGGAAACGACCCCGCUGAACCAGAUCCUCGGUGGGUACUUGCGGUCCGAGGUGAAGUGUCUGUCCUGCCAGC .......(((.......((.((((((....(((((((((((((.....(....)....(((((((.........))))))))))))...)))))..)))....)).)))).)))))... ( -35.41, z-score = 0.79, R) >apiMel3.Group1 24390819 116 - 25854376 UUAUUUAGCAAGA---CACAAAGCUAGUAAACUAGACAGUUAUUCUAAAGAAACAACACCUAUUAAUCAAAUUUUUGGUGGUUAUAUUCGUACUGAAGUGAAAUGUUUACAGUGUCGGC ...........((---(((....((((....)))).((((..(((....)))....((((................))))...........))))..((((.....)))).)))))... ( -19.49, z-score = 0.09, R) >triCas2.ChLG3 25585079 119 - 32080666 CUAUUUAAGUCGAUUUAAAAACCAUUCCGAAUUUGAUUCAAAAGUGAAAGAGACCACACCUCUCAAUCAAAUACUAGGCGGAUAUUUAAGAUCGGCAGUGAGAUGUUUGAAGUGUGGUC ....((((((((...............)).))))))((((....))))...((((((((.((...(((...((((..((.(((.......))).)))))).)))....)).)))))))) ( -23.26, z-score = -0.19, R) >consensus GUAUCUGAUGCGGUUUCGCAACUACAAAGAGCUGGAUCAGCUGGUGAAGGAGACGACGCCGCUGAGUCAGAUCUUUGGUGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGCAACC .........(((..................(((.....)))................(((((((((.......)))))))))....)))(((((..((........))..))))).... (-15.01 = -14.27 + -0.73)

| Location | 5,765,253 – 5,765,359 |
|---|---|
| Length | 106 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.51422 |
| G+C content | 0.52525 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -16.57 |
| Energy contribution | -15.57 |
| Covariance contribution | -1.01 |
| Combinations/Pair | 2.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.553438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
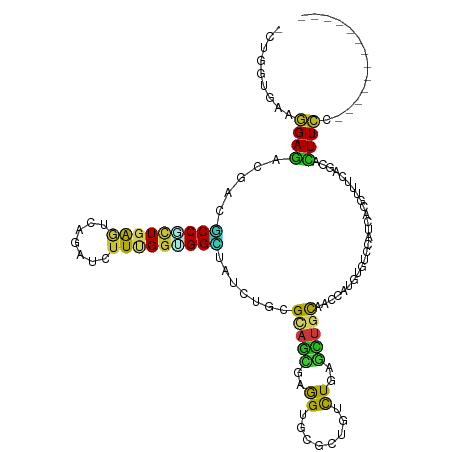
>dm3.chr3L 5765253 106 + 24543557 -UUGGUUAAGGAGACCACGCCGCUGGGACAGAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGCAACCAUGUGUCCAUUACGUUUCAGCACUUCC------------- -..(((...(....)...)))((((..((.........((((((....(.((((((.....)))))))..))))))......(((.....)))))..))))......------------- ( -35.80, z-score = -0.10, R) >droSim1.chr3L 5276996 106 + 22553184 -CUGGUAAAGGAGACCACGCCCCUGGGACAGAUCUUUGGGGGCUAUCUGCGCAGCGAGGUGCGGUGUCUGAGCUGCAACCAUGUGUCCAUCACGUUUCAGCACUUCC------------- -........(((.(((..(((((..(((....)).)..))))).....((((......))))))).)))..((((.(((...(((.....)))))).))))......------------- ( -35.30, z-score = 0.45, R) >droSec1.super_2 5702094 106 + 7591821 -CUGGUGAAGGAGACUACGCCGCUGGGACAGAUCUUUGGGGGCUAUCUGCGCAGCGAGGUGCGGUGCCUGAGCUGCAACCAUGUGUCCAUCACGUUUCAGCACUUCC------------- -..((((.((....)).))))((((..((.......(((..((.....))(((((.((((.....))))..)))))..))).(((.....)))))..))))......------------- ( -43.10, z-score = -1.40, R) >droEre2.scaffold_4784 8455858 106 + 25762168 -CUGGUAAAGGAGACUACGCCGCUGGGACAGAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGCCUGAGCUGCAACCAUGUGUCCAUCACGUUCCAGCACUUCC------------- -..(((..((....))..)))((((((((.........((((((....(.((((((.....)))))))..))))))......(((.....)))))))))))......------------- ( -44.60, z-score = -2.16, R) >droYak2.chr3L 6334752 106 + 24197627 -CUGGUAAAGGAGACUACGCCGCUGGGACAAAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGCAACCAUGUGUCCAUCACGUUCCAGCACUUCC------------- -..(((..((....))..)))((((((((.........((((((....(.((((((.....)))))))..))))))......(((.....)))))))))))......------------- ( -41.90, z-score = -2.00, R) >droAna3.scaffold_13337 10518092 106 - 23293914 -CUGGUUAAGGAGACGACGCCCCUGGGACAGAUCUUCGGCGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGUAACCAUGUGUCCAUCACGUUCCAGCACUUCC------------- -..(((...(....)...))).(((((((.........((((((....(.((((((.....)))))))..))))))......(((.....)))))))))).......------------- ( -33.40, z-score = 0.81, R) >dp4.chrXR_group6 3525682 106 - 13314419 -CUGGUGAAGGAGACGACCCCACUGAAUCAGAUAUUCGGUGGCUACCUGCGCAGCGAGGUGCGUUGCCUCAGCUGCAGCCACGUCUCAAUCACCUUCCAGCACUUCC------------- -..(((((..((((((...(((((((((.....)))))))))......((((((((((((.....))))).))))).))..))))))..))))).............------------- ( -47.10, z-score = -4.32, R) >droPer1.super_61 286113 106 - 350175 -CUGGUGAAGGAGACGACGCCACUGAAUCAGAUAUUCGGUGGCUACCUGCGCAGCGAGGUGCGUUGCCUCAGCUGCAGCCACGUCUCAAUCACCUUCCAGCACUUCC------------- -..(((((..((((((..((((((((((.....)))))))))).....((((((((((((.....))))).))))).))..))))))..))))).............------------- ( -51.30, z-score = -5.13, R) >droWil1.scaffold_180698 3794436 106 + 11422946 -UUGGUUAAGGAGACAACGCCGCUGAAUCAAAUAUUCGGUGGUUAUCUAAGAAGUGAAGUUCGUUGUUUGAGUUGUAAUCAUGUAUCGAUUACGUUUCAACAUUUCC------------- -........(((((((((((((((((((.....)))))))))).......(((......))))))).(((((.(((((((.......))))))).)))))..)))))------------- ( -28.50, z-score = -2.43, R) >droMoj3.scaffold_6680 11968746 106 + 24764193 -CUGGUGAAGGAGACGACGCCGCUGAGUCAAAUCUUUGGUGGCUAUUUGCGUAGCGAGGUUCGCUGCCUUAGCUGCAAUCAUGUCUCGAUUACGUUUCAACAUUUCC------------- -...((((..((((((..(((((..((.......))..))))).......((((((((((.....))))).))))).....))))))..))))..............------------- ( -34.50, z-score = -1.52, R) >droVir3.scaffold_13049 8314939 106 + 25233164 -CUGGUGAAGGAGACGACGCCAAUUAGUCAAAUCUUUGGCGGCUAUCUGCGCAGCGAGGUGCGCUGCCUCAGCUGCAAUCAUGUCUCGAUUACGUUUCAACAUUUCC------------- -...((((..((((((.((((((............))))))((.....))((((((((((.....))))).))))).....))))))..))))..............------------- ( -37.30, z-score = -2.07, R) >droGri2.scaffold_15110 7343463 106 + 24565398 -UUGGUGAAAGAGACGACGCCGUUGAAUCAAAUCUUUGGUGGCUAUCUGCGCAGCGAGGUGCGCUGCCUGAGCUGUAAUCAUGUCUCGAUAACGUUUCAACAUUUCC------------- -...((....((((((.(((((..((......))..)))))(((....(.((((((.....)))))))..)))........))))))....))..............------------- ( -30.60, z-score = -0.51, R) >anoGam1.chr2L 2226390 106 - 48795086 -UACAGCAAGGAAACGACCCCGCUGAACCAGAUCCUCGGUGGGUACUUGCGGUCCGAGGUGAAGUGUCUGUCCUGCCAGCACGUGUCGACCACGUUCCAGCACUUCG------------- -....(((((....)...((((((((.........))))))))...))))((..((.(((((.(((.(((......))))))...)).))).))..)).........------------- ( -33.90, z-score = -0.06, R) >apiMel3.Group1 24390855 120 - 25854376 UUAUUCUAAAGAAACAACACCUAUUAAUCAAAUUUUUGGUGGUUAUAUUCGUACUGAAGUGAAAUGUUUACAGUGUCGGCAUGUUUCUACAACUUUUCAACAUUUUCAAGUAUAUAUAAU .........(((((((.((((................)))).......((((((((.((........)).))))).)))..)))))))................................ ( -16.69, z-score = 0.19, R) >triCas2.ChLG3 25585119 106 - 32080666 -AAAGUGAAAGAGACCACACCUCUCAAUCAAAUACUAGGCGGAUAUUUAAGAUCGGCAGUGAGAUGUUUGAAGUGUGGUCACGUCAGCACCACCUUCCAACACUUUC------------- -((((((.....((((((((.((...(((...((((..((.(((.......))).)))))).)))....)).))))))))..(....)............)))))).------------- ( -26.60, z-score = -1.19, R) >consensus _CUGGUGAAGGAGACGACGCCGCUGAGUCAGAUCUUUGGUGGCUAUCUGCGCAGCGAGGUGCGCUGUCUGAGCUGCAACCAUGUGUCCAUCACGUUUCAGCACUUCC_____________ .........((((.....(((((((((.......))))))))).......(((((..((........))..)))))..........................)))).............. (-16.57 = -15.57 + -1.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:56 2011