
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,763,142 – 5,763,265 |
| Length | 123 |
| Max. P | 0.856095 |

| Location | 5,763,142 – 5,763,232 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.29 |
| Shannon entropy | 0.36544 |
| G+C content | 0.30046 |
| Mean single sequence MFE | -17.36 |
| Consensus MFE | -11.06 |
| Energy contribution | -9.96 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
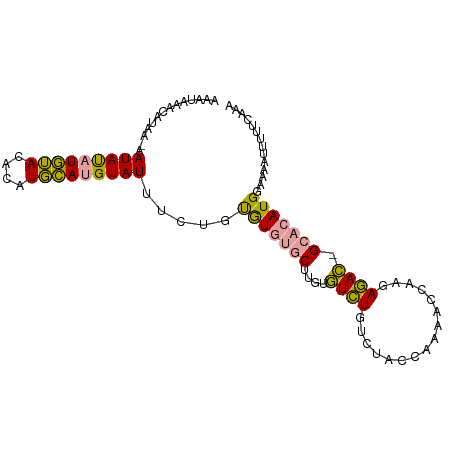
>dm3.chr3L 5763142 90 - 24543557 AAACAAACGGGACGAUACAUAAAUACAUUUAAGUAUAUAUACAU---CUUAUAUAUAU-----AAUAUGAAGAUAUAGCACAUGGAAAAUUGCGCAAA .........................(((....(((((((((...---..)))))))))-----...)))........((.((........)).))... ( -9.20, z-score = -0.63, R) >droPer1.super_61 284153 96 + 350175 AAAUAAACAUAA-AAUAUAUGUACACAUGCAUGUAUUUCUGUGUGUGCUUGUGUCUGUCUACCAAAACCAAGAGAC-GCACAUGGAAAAUUUUUCAAA ......((((((-((((((((((....))))))))))).)))))((((..((.(((..............))).))-)))).(((((....))))).. ( -21.44, z-score = -1.82, R) >dp4.chrXR_group6 3523966 96 + 13314419 AAAUAAACAUAA-AAUAUAUGUACACAUGCAUGUAUUUCUGUGUGUGCUUGUGUCUGUCUACCAAAACCAAGAGAC-GCACAUGGAAAAUUUUUCAAA ......((((((-((((((((((....))))))))))).)))))((((..((.(((..............))).))-)))).(((((....))))).. ( -21.44, z-score = -1.82, R) >consensus AAAUAAACAUAA_AAUAUAUGUACACAUGCAUGUAUUUCUGUGUGUGCUUGUGUCUGUCUACCAAAACCAAGAGAC_GCACAUGGAAAAUUUUUCAAA ..............(((((((((....))))))))).....(((((((....((((................)))).))))))).............. (-11.06 = -9.96 + -1.10)


| Location | 5,763,172 – 5,763,265 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 68.20 |
| Shannon entropy | 0.43045 |
| G+C content | 0.33869 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -9.44 |
| Energy contribution | -8.57 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5763172 93 - 24543557 UUUGUGGAGGGAUAUUCCCAAAAGAUUUGGCAAAAACAAACGGGACGAUACAUAAAUACAUUUAAGUAUAUAUACAU---CUUAUAUAUAUAAUAU ((((((..(((.....)))......((((.......))))..........)))))).........(((((((((...---..)))))))))..... ( -14.30, z-score = -1.20, R) >droPer1.super_61 284188 91 + 350175 -UUCCCUCUGACCAGGCC--AUAGUUUUGGCUAAAAUAAACAUAA-AAUAUAUGUACACAUGCAUGUAUUUCUGUGUGUGCUUGUGUCUGU-CUAC -........(((((((((--.((((....))))......((((((-((((((((((....))))))))))).)))))).))))).)))...-.... ( -23.70, z-score = -2.78, R) >dp4.chrXR_group6 3524001 91 + 13314419 -UUCCCUCUGACCAGGCC--AUAGUUUUGGCUAAAAUAAACAUAA-AAUAUAUGUACACAUGCAUGUAUUUCUGUGUGUGCUUGUGUCUGU-CUAC -........(((((((((--.((((....))))......((((((-((((((((((....))))))))))).)))))).))))).)))...-.... ( -23.70, z-score = -2.78, R) >consensus _UUCCCUCUGACCAGGCC__AUAGUUUUGGCUAAAAUAAACAUAA_AAUAUAUGUACACAUGCAUGUAUUUCUGUGUGUGCUUGUGUCUGU_CUAC ............(((((......(((((....)))))..(((((...(((((((((....)))))))))...)))))........)))))...... ( -9.44 = -8.57 + -0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:54 2011