| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,757,422 – 5,757,499 |
| Length | 77 |
| Max. P | 0.615008 |

| Location | 5,757,422 – 5,757,499 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Shannon entropy | 0.63737 |
| G+C content | 0.47069 |
| Mean single sequence MFE | -21.21 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
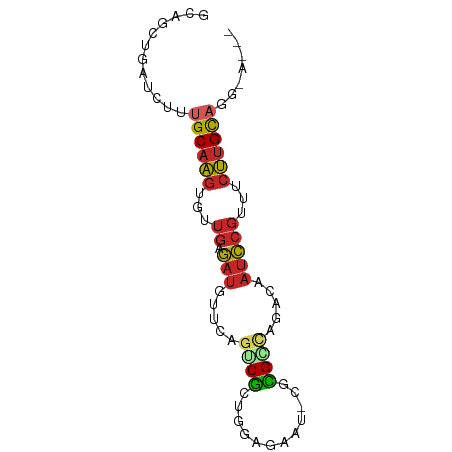
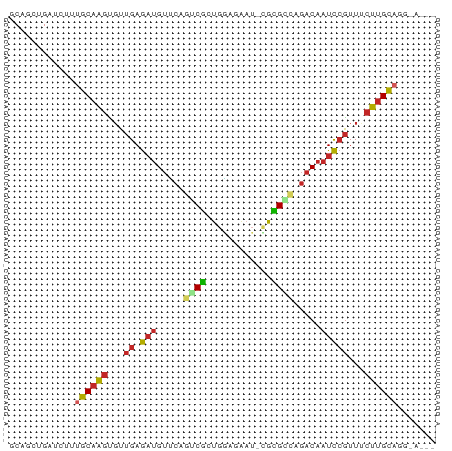
>dm3.chr3L 5757422 77 + 24543557 ACAGCUGAUCUUUGCAAGUGUUGAGAUGUUCAGUCGCUGGAGAAU-CGCGGCAGACAAUCCGUUUCUUGCAGGGAUUA .....(((((((((((((...((.((((((..(((((........-.))))).))).)))))...))))))))))))) ( -26.00, z-score = -1.73, R) >droAna3.scaffold_13337 10510137 72 - 23293914 GCAGAUGAUCUCUGCAAGCGUUGAGAUGUCCAGUCGCUGGAGAAU-CGUGACAGACAAUCCGUUUCUUGCAUG----- (((((.....)))))..(((..(((((((((((...)))))...(-(......)).....)))))).)))...----- ( -18.70, z-score = 0.15, R) >droYak2.chr3L 6326857 77 + 24197627 GCAGCUGAUCUUUGCAAGUGUUGAGAUGUUCAGUCGCUGGAGAAU-CGCGCCAGACAAUCCGUUUCUUGCAAGGACUA ........((((((((((...((.((((((..(.(((........-.))))..))).)))))...))))))))))... ( -20.70, z-score = 0.15, R) >droEre2.scaffold_4784 8447991 77 + 25762168 GCAGCUGAUCUUUGCAAGUGUUGAGAUGUUCAGUCGCUGGAUAAU-CGCGUCAGACAAUCCGUUUCUUGCAAGGAUUA .....(((((((((((((...((.((((((..(.(((........-.))).).))).)))))...))))))))))))) ( -23.10, z-score = -1.34, R) >droSec1.super_2 5694257 77 + 7591821 ACAGUUGAUCUAUGCAAGUGUUGAGAUGUUCAGUCGCUGGAAAAU-CGCGGCAGACAAUCCGUUUCUUGCAGGUAUUA .....((((...((((((...((.((((((..(((((........-.))))).))).)))))...))))))...)))) ( -18.20, z-score = -0.07, R) >droSim1.chr3L 5269183 77 + 22553184 ACAGUUGAUCUAUGCAAGUGUUGAGAUGUUCAGUCGCUGGAAAAU-CGCGGCAGACAAUCCGUUUCUUGCAGGUAUUA .....((((...((((((...((.((((((..(((((........-.))))).))).)))))...))))))...)))) ( -18.20, z-score = -0.07, R) >droWil1.scaffold_180698 4030744 73 + 11422946 UCCUUUGAUCUUUGCCGGACAUGAA--GCAAUAACUGUGUGGGAAAUAGGCCACACAAUUAUUUGGCAGUUGUUA--- .....((((..((((((........--..(((((.(((((((........))))))).)))))))))))..))))--- ( -19.20, z-score = -1.24, R) >droGri2.scaffold_15110 11173114 73 + 24565398 GCCAGUGCUUUGUGCAAGUGUUGAGAUGUGCUGGCGAUGUUUGAUAGACGCAAGACAAUCCGUUGCUUGCAAC----- (((((..(.((..((....))...)).)..)))))...(((.....)))(((((.(((....))))))))...----- ( -22.80, z-score = -1.15, R) >droVir3.scaffold_13049 11714577 75 + 25233164 GCCUGUGCUUUGCGCAGGUGUUGAGAUGUGCUGGCGCUGUACAUUAAACGCUAGACAAUCCGUUGCCUGCAUCCC--- ((((((((...))))))))(((..(((((((.......))))))).)))((.((.(((....))).)))).....--- ( -23.80, z-score = -0.95, R) >droMoj3.scaffold_6680 15655414 72 - 24764193 GCUAAUGAUCUAUGCAAGUGUUGAGAUGUGCUGGCGCUGUGUUUAAUACGCUAGACAAUCCGUUUCUUGCAG------ ............((((((...((.(((...((((((.(((.....)))))))))...)))))...)))))).------ ( -21.40, z-score = -1.76, R) >consensus GCAGCUGAUCUUUGCAAGUGUUGAGAUGUUCAGUCGCUGGAGAAU_CGCGCCAGACAAUCCGUUUCUUGCAGG_A___ ............((((((...((.(((.....((((............)))).....)))))...))))))....... (-10.09 = -10.15 + 0.06)

| Location | 5,757,422 – 5,757,499 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Shannon entropy | 0.63737 |
| G+C content | 0.47069 |
| Mean single sequence MFE | -17.04 |
| Consensus MFE | -8.07 |
| Energy contribution | -7.92 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.615008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
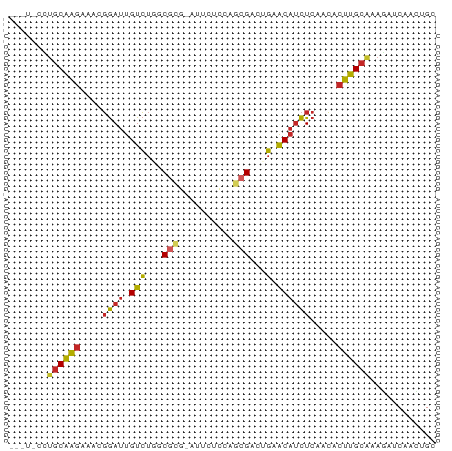
>dm3.chr3L 5757422 77 - 24543557 UAAUCCCUGCAAGAAACGGAUUGUCUGCCGCG-AUUCUCCAGCGACUGAACAUCUCAACACUUGCAAAGAUCAGCUGU ..(((..((((((....((((..((.(.(((.-........))).).))..)))).....))))))..)))....... ( -14.90, z-score = -0.21, R) >droAna3.scaffold_13337 10510137 72 + 23293914 -----CAUGCAAGAAACGGAUUGUCUGUCACG-AUUCUCCAGCGACUGGACAUCUCAACGCUUGCAGAGAUCAUCUGC -----..((((((....((((.(((((((.(.-........).))).)))))))).....))))))............ ( -17.70, z-score = -0.24, R) >droYak2.chr3L 6326857 77 - 24197627 UAGUCCUUGCAAGAAACGGAUUGUCUGGCGCG-AUUCUCCAGCGACUGAACAUCUCAACACUUGCAAAGAUCAGCUGC ..(((.(((((((....((((.((..(((((.-........))).))..)))))).....))))))).)))....... ( -19.10, z-score = -0.54, R) >droEre2.scaffold_4784 8447991 77 - 25762168 UAAUCCUUGCAAGAAACGGAUUGUCUGACGCG-AUUAUCCAGCGACUGAACAUCUCAACACUUGCAAAGAUCAGCUGC ..(((.(((((((....((((.(((......)-)).))))......(((.....)))...))))))).)))....... ( -17.50, z-score = -1.33, R) >droSec1.super_2 5694257 77 - 7591821 UAAUACCUGCAAGAAACGGAUUGUCUGCCGCG-AUUUUCCAGCGACUGAACAUCUCAACACUUGCAUAGAUCAACUGU .......((((((....(((..(((......)-))..)))......(((.....)))...))))))............ ( -14.30, z-score = -0.73, R) >droSim1.chr3L 5269183 77 - 22553184 UAAUACCUGCAAGAAACGGAUUGUCUGCCGCG-AUUUUCCAGCGACUGAACAUCUCAACACUUGCAUAGAUCAACUGU .......((((((....(((..(((......)-))..)))......(((.....)))...))))))............ ( -14.30, z-score = -0.73, R) >droWil1.scaffold_180698 4030744 73 - 11422946 ---UAACAACUGCCAAAUAAUUGUGUGGCCUAUUUCCCACACAGUUAUUGC--UUCAUGUCCGGCAAAGAUCAAAGGA ---.......((((.(((((((((((((........)))))))))))))((--.....))..))))............ ( -21.10, z-score = -2.98, R) >droGri2.scaffold_15110 11173114 73 - 24565398 -----GUUGCAAGCAACGGAUUGUCUUGCGUCUAUCAAACAUCGCCAGCACAUCUCAACACUUGCACAAAGCACUGGC -----...((((((((....))).)))))..............(((((..............(((.....)))))))) ( -14.13, z-score = 0.23, R) >droVir3.scaffold_13049 11714577 75 - 25233164 ---GGGAUGCAGGCAACGGAUUGUCUAGCGUUUAAUGUACAGCGCCAGCACAUCUCAACACCUGCGCAAAGCACAGGC ---((((((.((((((....)))))).(((((........))))).....))))))....((((.((...)).)))). ( -22.20, z-score = -0.93, R) >droMoj3.scaffold_6680 15655414 72 + 24764193 ------CUGCAAGAAACGGAUUGUCUAGCGUAUUAAACACAGCGCCAGCACAUCUCAACACUUGCAUAGAUCAUUAGC ------.((((((....((((.((((.((((..........)))).)).)))))).....))))))............ ( -15.20, z-score = -1.27, R) >consensus ___U_CCUGCAAGAAACGGAUUGUCUGGCGCG_AUUCUCCAGCGACUGAACAUCUCAACACUUGCAAAGAUCAACUGC .......((((((....((((.(((...(((..........)))...).)))))).....))))))............ ( -8.07 = -7.92 + -0.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:51 2011