
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,757,109 – 5,757,183 |
| Length | 74 |
| Max. P | 0.679115 |

| Location | 5,757,109 – 5,757,183 |
|---|---|
| Length | 74 |
| Sequences | 11 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 71.65 |
| Shannon entropy | 0.56057 |
| G+C content | 0.49938 |
| Mean single sequence MFE | -22.29 |
| Consensus MFE | -13.10 |
| Energy contribution | -12.89 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
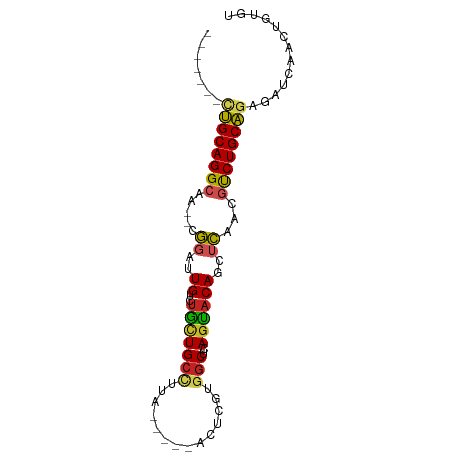
>dm3.chr3L 5757109 74 - 24543557 -------CUGCAGGCAA--CGGAUUGUCUGCUGCCUUA-----ACUCGUGGCUCAGCACAGCUCAACGUCUGCAGAGAUCAACAGUGU -------((((((((..--.(..(((((((..(((...-----......))).))).))))..)...))))))))............. ( -24.30, z-score = -0.75, R) >droSim1.chr3L 5268869 74 - 22553184 -------UUGCAGGCAA--CGGAUUGUCUGUUGCCUUA-----ACUCGUGGCUCAGCACAGCUCAACGUCUGCAGAGAUCAACAGUGU -------((((((((..--.(..(((((((..(((...-----......))).))).))))..)...))))))))............. ( -22.30, z-score = -0.51, R) >droSec1.super_2 5693943 74 - 7591821 -------UUGCAGGCAA--CGGAUUGUCUGUUGCCUUA-----ACUCGUGGCUCAGCACAGCUCAACGUCUGCAGAGAUCAACUGUGU -------((((((((..--.(..(((((((..(((...-----......))).))).))))..)...))))))))............. ( -22.30, z-score = -0.44, R) >droEre2.scaffold_4784 8447678 74 - 25762168 -------CUGCAGGCAA--CGGAUUGUCUGCUGCCCUU-----ACUCGUGGCUCAGUACAGCUCAAAGUCUGCAGAGAUGAACAGUGU -------((((((((..--.(..(((((((..((((..-----....).))).))).))))..)...))))))))............. ( -24.20, z-score = -0.71, R) >droYak2.chr3L 6326545 74 - 24197627 -------CUGCAGGCAA--CGGAUUGUCUGCUGCCUUA-----ACUCGUGGCUCAGUACAGCUCAAAGUCUGCAGAGAUCAACAGUGU -------((((((((..--.(..(((((((..(((...-----......))).))).))))..)...))))))))............. ( -24.30, z-score = -1.14, R) >droAna3.scaffold_13337 10509829 73 + 23293914 -------UCGCAGGCAA--CGGAUUGUCUAAUGCC-GA-----ACUCCUGGCUUAGUUCAUCUCAAAGUCUGCGGAGAAUAGAUGCGU -------((((((((..--.((((...((((.(((-(.-----.....))))))))...))))....))))))))............. ( -21.80, z-score = -1.29, R) >dp4.chrXR_group6 3517852 74 + 13314419 -------CUGCAGGCAAAGCUAAAUGGCUGCUGCUUUACCAUU-----UUGCACAGUACACCCAAACUCCUGCAGAUAUCAAUUGU-- -------(((((((............((((.(((.........-----..)))))))...........)))))))...........-- ( -18.70, z-score = -1.02, R) >droPer1.super_61 278037 74 + 350175 -------CUGCAGGCAAAGCUAAAUGGCUGCUGCUUUACCAUU-----UUGCACAGUACACCCAAACUCCUGCAGAUAUCAAUUGU-- -------(((((((............((((.(((.........-----..)))))))...........)))))))...........-- ( -18.70, z-score = -1.02, R) >droVir3.scaffold_13049 11714389 86 - 25233164 CCUUUUGCUGCAGGCAA--CGGAAUGGCUGUUGACCCUAAAUUAUAACUGGCACAGCACAGCUCAACGUCUGCAUAGAUCAAUCGCGG .....((((((((((..--..((.(((((((.(.((.............)))))))).))..))...)))))))..((....))))). ( -20.42, z-score = 0.05, R) >droMoj3.scaffold_6680 15655248 86 + 24764193 CCUUCUGCUGCAGGCAA--CGGAUUGGCUGCUGCCCAACAAAUAUAAAUGGCACAGUACAGCUCAACGUCUGCAGAGAUCAAUUGCGG ...(((.((((((((..--.(..(((((((.((((..............)))))))).)))..)...))))))))))).......... ( -26.34, z-score = -1.19, R) >droGri2.scaffold_15110 11172940 86 - 24565398 CCAUCUGCUGCAGGCAA--CGGAUUGUCUGCUGCUUUAUAAAUACACCUGGCACAGCACAGCUCAACGUCUGCAUAAAUCAAUUGCGG ....(((((((((((..--.(..(((((((.((((..............))))))).))))..)...)))))))..........)))) ( -21.84, z-score = -0.63, R) >consensus _______CUGCAGGCAA__CGGAUUGUCUGCUGCCUUA_____ACUCGUGGCUCAGUACAGCUCAACGUCUGCAGAGAUCAACUGUGU .......((((((((.........((.(((..(((..............))).))))).........))))))))............. (-13.10 = -12.89 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:49 2011