| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,756,483 – 5,756,575 |
| Length | 92 |
| Max. P | 0.998406 |

| Location | 5,756,483 – 5,756,575 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 62.49 |
| Shannon entropy | 0.74812 |
| G+C content | 0.52335 |
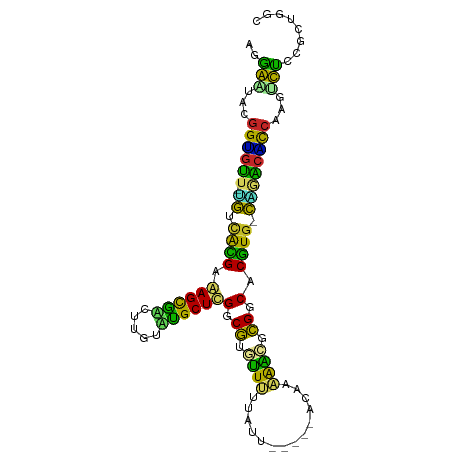
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -23.33 |
| Energy contribution | -18.79 |
| Covariance contribution | -4.54 |
| Combinations/Pair | 2.18 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
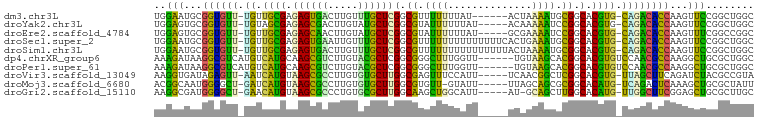
>dm3.chr3L 5756483 92 - 24543557 UGGAAUGCGGUGUU-UGUUGCGAGAGUGACUUGUUUGCUCGGCGUUUUUUUAU------ACUAAAAUGCGGCACGUG-CAGACACCAAGUUCCGGCUGGC .(((((..((((((-((((((..(((..(.....)..))).(((((((.....------...))))))).)))...)-))))))))..)))))....... ( -33.80, z-score = -2.35, R) >droYak2.chr3L 6325941 93 - 24197627 UGGAGUGCGGUGUU-UGUAGCGAGAGCGACUUGUAUGCUCGGCGUAUUUUUUAU-----ACAAAAAUCCGGCACGUG-CAGACACCAAGUUCCGGCUGGC .(((((..((((((-((((.((.(((((.......)))))(.((.((((((...-----..)))))).)).).))))-))))))))..)))))....... ( -30.20, z-score = -0.89, R) >droEre2.scaffold_4784 8447060 93 - 25762168 UGGAGUGCGGUGUU-UGUUGCGAGAGCAACUUGUAUGCUCGGCGUAUUUUUUAU-----GCGAAAAUCCGGCACGUG-CAGACACCAAGUUUCGGCCGGC .(((((..((((((-((((((....)))))..(((((.(((((((((.....))-----))).....))))..))))-))))))))..)))))....... ( -28.90, z-score = 0.03, R) >droSec1.super_2 5693382 98 - 7591821 UGGAAUGCGGUGUU-UGUUGCGAGAGUGAAUUGUUUGCUCGGCGUUUUUUUUUUUUUUCACUGAAAUGCGGCACGUG-CAGACACCAAGUUCCGGCUGGC .(((((..((((((-((((((..(((..(.....)..))).(((((((..............))))))).)))...)-))))))))..)))))....... ( -33.04, z-score = -1.71, R) >droSim1.chr3L 5268238 98 - 22553184 UGGAAUGCGGUGUU-UGUUGCGAGAGUGACUUGUUUGCUCGGCGUUUUUUUUUUUUUUUACUAAAAUGCGGCACGUG-CAGACACCAAGUUCCGGCUGGC .(((((..((((((-((((((..(((..(.....)..))).(((((((..............))))))).)))...)-))))))))..)))))....... ( -32.54, z-score = -1.94, R) >dp4.chrXR_group6 3517329 94 + 13314419 AAAGAUAAGGCGUCAUGUCAUGCAAGCGUCUUGUACGCUCGGCGGGCUUUGGUU------UGUAAGCACGGCACGUGUCCAACGCCAAGGCUGCGCUGGC ....(((((((((..((.....)).)))))))))..((.(((((((((((((((------((...(((((...))))).))).))))))))).))))))) ( -37.10, z-score = -1.34, R) >droPer1.super_61 277514 94 + 350175 AAAGAUAAGGCGUCAUGUCAUGCAAGCGUCUUGUACGCUCGGCGGGCUUUGGUU------UGUAAGCACGGCACGUGUCCAACGCCAAGGCUGCGCUGGC ....(((((((((..((.....)).)))))))))..((.(((((((((((((((------((...(((((...))))).))).))))))))).))))))) ( -37.10, z-score = -1.34, R) >droVir3.scaffold_13049 11713847 93 - 25233164 AAGGUGAUAGAGUU-AAUCAUGUAAGCGCCUUGUGUGCUUGGCGAGUUUCCAUU-----UCAACGGCUCGGCACGUG-UUAGCUUCAGAUCUACGCCGUA ..((((.(((((((-((.(((((((((((.....))))))(.((((((......-----.....)))))).))))))-)))))......))))))))... ( -28.20, z-score = -1.07, R) >droMoj3.scaffold_6680 15654688 92 + 24764193 ACGGCAAUGGGGCU-GAUCAUGUAAGCGCCUUGUGUGCUUGGCGUGUU-GUAUU-----UUAGCAGCGCGGCACAUG-UCAGACUCAAAGCUGCGCUAUU .((((..((((.((-((.(((((((((((.....))))))(.((((((-((...-----...)))))))).))))))-)))).))))..))))....... ( -38.30, z-score = -2.21, R) >droGri2.scaffold_15110 11172386 92 - 24565398 AAGGCGAUGGGGCU-GAACAUGUAAGCGCCCUGUGCGCUUGGCAAGCUGGCAUU-----AU-GCAGCUUGGCACAUG-UUGGCUUCGGAGCUGCGCUUGC ..(((..(((((((-.(((((((((((((.....))))))(.(((((((.....-----..-.))))))).))))))-)))))))))..)))........ ( -41.00, z-score = -1.43, R) >consensus AGGAAUACGGUGUU_UGUCACGAAAGCGACUUGUAUGCUCGGCGUGUUUUUAUU_____ACAAAAACGCGGCACGUG_CAGACACCAAGUCUCCGCUGGC ..(((...((((((.((.((((.((((((.....))))))(.((.((((..............)))).)).).)))).))))))))...)))........ (-23.33 = -18.79 + -4.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:49 2011