| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,735,091 – 5,735,196 |
| Length | 105 |
| Max. P | 0.943331 |

| Location | 5,735,091 – 5,735,196 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.84 |
| Shannon entropy | 0.30127 |
| G+C content | 0.59363 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.30 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
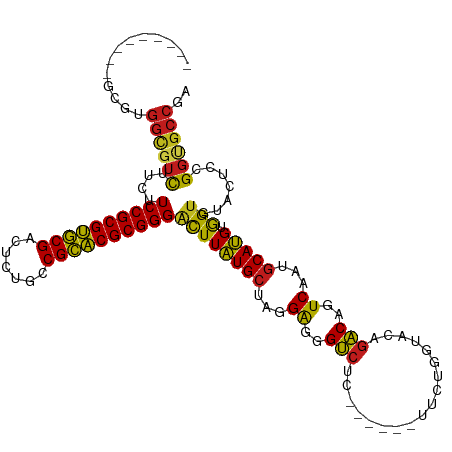
>dm3.chr3L 5735091 105 + 24543557 UCGGCACCGGAGUACACUACAUGCAUCGACUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCACGU--------- ..(((...............((((...((..((((.(......)------.))))..))...))))..((((((((((((((...))))))))))).....))))))....--------- ( -36.40, z-score = -1.68, R) >droSim1.chr3L 5247053 105 + 22553184 UCGGCACCGGAGUACACCACAUGCAUCGACUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACACCACGC--------- ..(((...((......))..((((...((..((((.(......)------.))))..))...))))..)))(((((((((((...)))))))))))...............--------- ( -32.60, z-score = -1.02, R) >droSec1.super_2 5672193 105 + 7591821 UCGGCACCGGAGUACACCACAUGCAUCGACUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCACGC--------- ..(((...((......))..((((...((..((((.(......)------.))))..))...))))..((((((((((((((...))))))))))).....))))))....--------- ( -37.00, z-score = -1.90, R) >droYak2.chr3L 6304325 105 + 24197627 UCGGCACCGGAGUACACCACAUGCAUCGACUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCACGC--------- ..(((...((......))..((((...((..((((.(......)------.))))..))...))))..((((((((((((((...))))))))))).....))))))....--------- ( -37.00, z-score = -1.90, R) >droEre2.scaffold_4784 8426121 105 + 25762168 UCGGCACCGGAGUACACCACAUGCAUCGACUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCACGC--------- ..(((...((......))..((((...((..((((.(......)------.))))..))...))))..((((((((((((((...))))))))))).....))))))....--------- ( -37.00, z-score = -1.90, R) >droAna3.scaffold_13337 10486331 105 - 23293914 UCGGCCCCGGAGUACACCACAUGCAUUGAUUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCCCGG--------- ......((((.((.......((((...((..((((.(......)------.))))..))...))))..((((((((((((((...))))))))))).....))))).))))--------- ( -37.70, z-score = -2.48, R) >dp4.chrXR_group6 5424545 105 - 13314419 UCGGCGCCCGAGUACACCACAUGCAUUGACUGUCUGUACCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAAAGUCGCACGCGGAAGAAAACGCCCCGA--------- ((((.((.............((((...((..((((.(......)------.))))..))...)))).....(((((((((((...)))))))))))........)).))))--------- ( -36.20, z-score = -2.81, R) >droPer1.super_27 571658 105 + 1201981 UCGGCGCCCGAGUACACCACAUGCAUUGACUGUCUGUACCAGAA------GCGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAAAGUCGCACGCGGAAGAAAACGCCCCGA--------- ((((.((.............((((...((..(((.((.......------)))))..))...)))).....(((((((((((...)))))))))))........)).))))--------- ( -34.30, z-score = -1.77, R) >droWil1.scaffold_180698 4002421 105 + 11422946 UCGGCUCCCGAGUAUACCACAUGCAUUGAUUGUCUCUAUCAGAA------GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAAAGUCGCACGCGGAAGAAGACACCAAGA--------- ((((...)))).........((((...((..((((((......)------)))))..))...))))..((((((((((((((...))))))))))).....))).......--------- ( -37.10, z-score = -3.86, R) >droVir3.scaffold_13049 11689578 114 + 25233164 UCGGCGCCCGAAUACACCACAUGCAUUGACUGUCUGUACCAGAA------GCGACCCUCCUAGCAUAAGCCCCGCGUGCGGCAAAGUCGCACGCGGAAGAAGACGCCACGCGGCCUAAGC ..((.(((((........(((.(((.....))).))).......------(((.........((....)).(((((((((((...))))))))))).......)))..)).))))).... ( -35.50, z-score = -0.60, R) >droMoj3.scaffold_6680 15631756 114 - 24764193 UCGGCGCCCGAGUACACCACAUGCAUUGACUGUCUGUACCAGAA------GCGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCACGCGGACUGAGC ..(((((.(..(((((..(((.(......)))).)))))..)..------))).))......((...(((((((((((((((...))))))))))).......(((...)))))))..)) ( -37.20, z-score = -0.19, R) >droGri2.scaffold_15110 11150659 105 + 24565398 UCGGCACCAGAAUAUACCACAUGCAUUGAUUGUCUGUAUCAGAA------ACGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAAAGUCGCACGCGGAAGAAGACGCCACGU--------- ..(((...............((((...((..(((.((.......------)))))..))...))))..((((((((((((((...))))))))))).....))))))....--------- ( -34.20, z-score = -2.74, R) >anoGam1.chr2L 15069742 111 - 48795086 UCCCAGUCGACGCACGCUUCUUGUAUGGAGUGCAUCAUGCAGGACAACAUGAAGCCUCCAUAGCAAAAAGCCCGCGCACGGCAGAGCCGUGCGCGGAAGAAGUCGUCCCGG--------- .((..(.((((....((((.((((((((((.((.(((((........))))).)))))))).)))).))))(((((((((((...))))))))))).....)))).)..))--------- ( -55.50, z-score = -6.44, R) >consensus UCGGCACCGGAGUACACCACAUGCAUUGACUGUCUGUACCAGAA______GAGACCCUCCUAGCAUAAGUCCCGCGUGCGGCAGAGUCGCACGCGGAAGAAGACGCCACGC_________ ..((................((((...((..(((..................)))..))...)))).....(((((((((((...))))))))))).........))............. (-26.70 = -26.30 + -0.40)


| Location | 5,735,091 – 5,735,196 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.84 |
| Shannon entropy | 0.30127 |
| G+C content | 0.59363 |
| Mean single sequence MFE | -45.72 |
| Consensus MFE | -35.82 |
| Energy contribution | -35.19 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5735091 105 - 24543557 ---------ACGUGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAGUCGAUGCAUGUAGUGUACUCCGGUGCCGA ---------(((((.((((.....(((((((((.......)))))))))((((..(((((((((.....)------)))))))).....))))))))))))).(.((((....))))).. ( -43.40, z-score = -1.50, R) >droSim1.chr3L 5247053 105 - 22553184 ---------GCGUGGUGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAGUCGAUGCAUGUGGUGUACUCCGGUGCCGA ---------((((.((((((..(((((((((((.......)))))))))))....(((((((((.....)------)))))))).)))))..).))))...(((..(......)..))). ( -43.20, z-score = -1.20, R) >droSec1.super_2 5672193 105 - 7591821 ---------GCGUGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAGUCGAUGCAUGUGGUGUACUCCGGUGCCGA ---------(((((((((((..(((((((((((.......)))))))))))....(((((((((.....)------)))))))).)))).))).))))...(((..(......)..))). ( -44.30, z-score = -1.30, R) >droYak2.chr3L 6304325 105 - 24197627 ---------GCGUGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAGUCGAUGCAUGUGGUGUACUCCGGUGCCGA ---------(((((((((((..(((((((((((.......)))))))))))....(((((((((.....)------)))))))).)))).))).))))...(((..(......)..))). ( -44.30, z-score = -1.30, R) >droEre2.scaffold_4784 8426121 105 - 25762168 ---------GCGUGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAGUCGAUGCAUGUGGUGUACUCCGGUGCCGA ---------(((((((((((..(((((((((((.......)))))))))))....(((((((((.....)------)))))))).)))).))).))))...(((..(......)..))). ( -44.30, z-score = -1.30, R) >droAna3.scaffold_13337 10486331 105 + 23293914 ---------CCGGGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAAUCAAUGCAUGUGGUGUACUCCGGGGCCGA ---------((((((.((((..(((((((((((.......)))))))))))....(((((((((.....)------)))))))).))))......(((((...)))))))))))...... ( -44.90, z-score = -1.95, R) >dp4.chrXR_group6 5424545 105 + 13314419 ---------UCGGGGCGUUUUCUUCCGCGUGCGACUUUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGGUACAGACAGUCAAUGCAUGUGGUGUACUCGGGCGCCGA ---------.....(((((.....(((((((((.......)))))))))((((..(((((((((.....)------)))))))).....)))))))))...((((((......)))))). ( -42.00, z-score = -1.42, R) >droPer1.super_27 571658 105 - 1201981 ---------UCGGGGCGUUUUCUUCCGCGUGCGACUUUGCCGCACGCGGGACUUAUGCUAGGAGGGUCGC------UUCUGGUACAGACAGUCAAUGCAUGUGGUGUACUCGGGCGCCGA ---------.....(((((.....(((((((((.......)))))))))((((..(((((((((.....)------)))))))).....)))))))))...((((((......)))))). ( -42.20, z-score = -1.11, R) >droWil1.scaffold_180698 4002421 105 - 11422946 ---------UCUUGGUGUCUUCUUCCGCGUGCGACUUUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC------UUCUGAUAGAGACAAUCAAUGCAUGUGGUAUACUCGGGAGCCGA ---------(((((((((((..(((((((((((.......)))))))))))..(((((...((..(((((------(......))))))..))...))))).)).)))).)))))..... ( -43.70, z-score = -3.27, R) >droVir3.scaffold_13049 11689578 114 - 25233164 GCUUAGGCCGCGUGGCGUCUUCUUCCGCGUGCGACUUUGCCGCACGCGGGGCUUAUGCUAGGAGGGUCGC------UUCUGGUACAGACAGUCAAUGCAUGUGGUGUAUUCGGGCGCCGA (((((.(((((((((((((((((((((((((((.......)))))))))(((....)))))))))).)))------..(((.......)))......)))))))).))....)))..... ( -48.50, z-score = -0.98, R) >droMoj3.scaffold_6680 15631756 114 + 24764193 GCUCAGUCCGCGUGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCGC------UUCUGGUACAGACAGUCAAUGCAUGUGGUGUACUCGGGCGCCGA ((((((((((((((.(((......(((((((((.......)))))))))((((..(((((((((.....)------)))))))).....)))).))))))))))...))).))))..... ( -48.70, z-score = -1.08, R) >droGri2.scaffold_15110 11150659 105 - 24565398 ---------ACGUGGCGUCUUCUUCCGCGUGCGACUUUGCCGCACGCGGGACUUAUGCUAGGAGGGUCGU------UUCUGAUACAGACAAUCAAUGCAUGUGGUAUAUUCUGGUGCCGA ---------...((((..(....((((((((((.......))))))))))((((((((...((..(((((------.......)).)))..))...))))).))).......)..)))). ( -38.70, z-score = -1.71, R) >anoGam1.chr2L 15069742 111 + 48795086 ---------CCGGGACGACUUCUUCCGCGCACGGCUCUGCCGUGCGCGGGCUUUUUGCUAUGGAGGCUUCAUGUUGUCCUGCAUGAUGCACUCCAUACAAGAAGCGUGCGUCGACUGGGA ---------((((..((((.....(((((((((((...)))))))))))((((((((.(((((((((.((((((......)))))).)).)))))))))))))))....)))).)))).. ( -66.10, z-score = -7.97, R) >consensus _________GCGUGGCGUCUUCUUCCGCGUGCGACUCUGCCGCACGCGGGACUUAUGCUAGGAGGGUCUC______UUCUGGUACAGACAGUCAAUGCAUGUGGUGUACUCCGGUGCCGA .............((((((....((((((((((.......))))))))))((((((((...((..(((..................)))..))...))))).))).......)))))).. (-35.82 = -35.19 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:48 2011