| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,407,971 – 4,408,090 |
| Length | 119 |
| Max. P | 0.710605 |

| Location | 4,407,971 – 4,408,090 |
|---|---|
| Length | 119 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.17 |
| Shannon entropy | 0.62226 |
| G+C content | 0.60824 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -17.37 |
| Energy contribution | -16.75 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
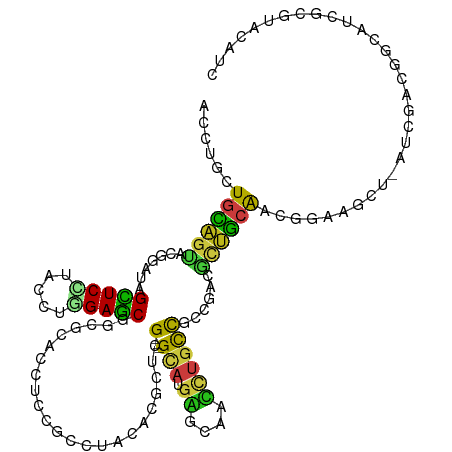
>dm3.chr2L 4407971 119 - 23011544 ACCUGCUGCAGUACCGAUAGCUCCUACCUGGAGCGGCGCACGUCUGCCUACAUGGGUGGUUGCAUGGGCAACCUGCCGCCCACGCUGCAACGGAAGCU-AUCGACUGCCUCGCGUACAUC ....((.(((((..((((((((((....((.(((((((......))))....((((((((.....((....)).)))))))).))).))..)).))))-)))))))))...))....... ( -50.30, z-score = -2.20, R) >droSim1.chr2L 4332395 119 - 22036055 ACCUGCUGCAGUACCGAUAGCUCCUACCUAGAGCGGCGCACGUCUGCCUACAUGGGCGGUUGCAUGGGCAACCUGCCGCCCACGCUGCAACGGAAGCU-GUCGACUGCCUCGCGUACAUC ....((.(((((..((((((((((........((((((((....))).....((((((((.....((....)).)))))))).)))))...)).))))-)))))))))...))....... ( -49.30, z-score = -1.80, R) >droSec1.super_5 2506257 119 - 5866729 ACCUGCUGCAGUACCGAUAGCUCCUACCUGGAGCGGCGCACGUCUGCCUACAUGGGCGGUUGCAUGGGCAACCUGCCGCCCACGCUGCAACGGAAGCU-GUCGACUGCUUCGCGUACAUC ....((.(((((..((((((((((....((.(((((((......))))....((((((((.....((....)).)))))))).))).))..)).))))-)))))))))...))....... ( -52.90, z-score = -2.57, R) >droYak2.chr2L 4441474 119 - 22324452 ACCUGCUGCAGUACCGAUAGCUCCUACUUGGAGCGGCGCACGUCUGCCUACAUGGGCGGUUGCAUGGGCAACCUGCCGCCCACGCUGCAACGGAAGCU-AUCAACUGCCUCGCGGACUUC ..((((.(((((...(((((((((...(((.(((((((......))))....((((((((.....((....)).)))))))).))).))).)).))))-))).)))))...))))..... ( -54.10, z-score = -3.57, R) >droEre2.scaffold_4929 4490451 119 - 26641161 ACCUGCUGCAGUACCGAUAGCUCCUACCUGGAGCGCCGCUCAUCUGCCUACAUGGGCGGUUGCAUGGGCAACCUGCCGCCUACGAUGCAACGGAAGCU-AUCGACUGCCUCGCGAACAUC ...(((.(((((..((((((((((.....))))).(((..((((.((((....))))(((.(((.((....))))).)))...))))...)))....)-)))))))))...)))...... ( -46.70, z-score = -2.69, R) >droAna3.scaffold_12943 1595123 119 + 5039921 ACGUGCUGUAGUACAGACAGCUCCUACCUAGAGCGGCGCACCUCCGCCUACAUGGGCGGCUGUAUGGGCAACCUGCCUCCGGCCUUAGACCGCAAAAU-GUCGACGGCUUCGCGAACGUC .((((..((.((...((((((((.......)))).(((.......((((....))))(((((...((((.....)))).)))))......)))....)-))).)).))..))))...... ( -40.20, z-score = 0.35, R) >droPer1.super_8 3429134 117 + 3966273 ACCUGCUGCAGCACGGACAGCUCUUACCUAGAGCGUCGCACCUCCGCCUAUACGCUGG---GCAUGAGCAAUCUGCCGCCGACGCUGCAGAGGAAGCUAAUCGACGGCAUCGCGCACAUC .(((.(((((((.......(((((.....)))))(((((......((......))..(---(((.((....))))))).))))))))))))))..(((.......)))............ ( -41.70, z-score = -0.76, R) >dp4.chr4_group3 2345504 116 + 11692001 ACCUGCUGCAGCACGGACAGCUCUUACCUAGAGCGUCGCACCUCCGCCUAUACGCUGG---GCAUGAGCAAUCUGCCGCCGACGCUGCAGAGGAAGCU-AUCGACGGCAUCGCGCACAUC .(((.(((((((.......(((((.....)))))(((((......((......))..(---(((.((....))))))).))))))))))))))..(((-......)))............ ( -42.20, z-score = -0.77, R) >droWil1.scaffold_180772 4795072 116 + 8906247 ACUUGCUGCAGCACGGACAGUUCGUAUCUGGAACGGCGCACUUCGGCCUAUACCUUGG---GCAUGAGCAAUUUACCACCUACAUUGCAACGGAAAUU-AUCGACAGCUUCGCGUACAUC ....(((((.((.((..(((.......)))...))))....(((((((((.....)))---))....(((((.((.....)).)))))..))))....-...).))))............ ( -26.10, z-score = 0.59, R) >droVir3.scaffold_12963 2423951 116 + 20206255 ACUUGUUGCAGCACGGAUAGCUCCUACCUGGAGCGUCGCACCUCGGCCUAUACGCUGG---GCAUGAGCAAUCUGCCGCCGACGCUGCAGCGUAAAUU-GUCGACGGCAUCGCGCACUUC ......((((((.......(((((.....)))))(((((....((((......))))(---(((.((....))))))).))))))))))((((....(-((.....)))..))))..... ( -45.70, z-score = -1.54, R) >droMoj3.scaffold_6500 9540761 116 + 32352404 ACCUGCUGCAGCACGGAUAGCUCCUAUCUGGAGCGUCGCACUUCCGCCUAUACGCUGG---GCAUGAGCAAUCUGCCACCGACGCUGCAGCGUAAGUU-GUCAACGGCAUCGCGCACAUC ....((((((((.......(((((.....)))))((((.......((......))..(---(((.((....))))))..))))))))))))((..(.(-((.....))).)..))..... ( -44.10, z-score = -1.52, R) >droGri2.scaffold_15252 1973710 116 + 17193109 ACCUGUUGCAGCACUGAUAGCUCCUAUCUGGAGCGACGGACCUCUGCCUACACGCUGG---GCAUGAGCAAUCUGCCGCCGACGCUGCAACGGAAGCU-GUCGACUGCGUCGCGUACAUC ..((((((((((.......(((((.....)))))(.((((.(((((((((.....)))---))).)))...)))).)......))))))))))..((.-(.((....)).)))....... ( -44.10, z-score = -1.73, R) >anoGam1.chr3R 44542973 104 + 53272125 ACAUGCUGCAGUACGGAGAGUUCCUUCCUCGAGCGGCGAUGCUCCGCCAUCACGCUCG---GCCUUUCCUCGCUACCACCGAUAUCGCGGUCACCGU--CGCGGCAGCA----------- ...((((((.....((((((........(((((((..((((......)))).))))))---).)))))).(((.((.((((......))))....))--.)))))))))----------- ( -37.70, z-score = -1.09, R) >triCas2.ChLG4 9167785 103 - 13894384 ACGUGCUCGUCUUCAGGCAGCUCUUACUUGGAGCGCCGUGAUUCCGCGGUGGCGAUAGG--GCUUCCGG----UGUUGCCGGGACACAAGUCGAAGA--AUCGUCGCCGUC--------- (((.((.(((((((.(((.(((((.....))))))))(((..(((.(((..(((...((--....))..----)))..))))))))).....)))))--..))..))))).--------- ( -41.90, z-score = -1.13, R) >consensus ACCUGCUGCAGUACGGAUAGCUCCUACCUGGAGCGGCGCACCUCCGCCUACACGCUCG___GCAUGAGCAACCUGCCGCCGACGCUGCAACGGAAGCU_AUCGACGGCAUCGCGUACAUC ......((((((.......(((((.....)))))...........................(((.((....))))).......))))))............................... (-17.37 = -16.75 + -0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:21 2011