| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,716,523 – 5,716,616 |
| Length | 93 |
| Max. P | 0.928541 |

| Location | 5,716,523 – 5,716,616 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 72.64 |
| Shannon entropy | 0.60944 |
| G+C content | 0.46490 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
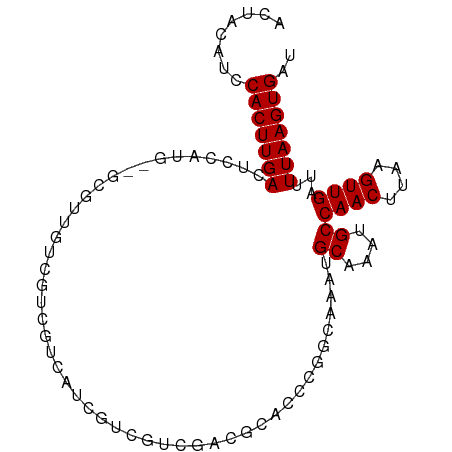
>dm3.chr3L 5716523 93 - 24543557 ACUACAUCCACUUGACUCCAUG--GCGUUGUCGUCGUCAUCGUCGUCGACGUACCCGGCAAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((.......--((((((.((.((....)).)).))))))....((((........))))..............))))))).. ( -24.60, z-score = -1.67, R) >droSim1.chr3L 5228836 93 - 22553184 ACUACAUCCACUUGACUCCAUG--GCGUUGUCGUCGUCAUCGUCGUCGACGUACCCGGCAAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((.......--((((((.((.((....)).)).))))))....((((........))))..............))))))).. ( -24.60, z-score = -1.67, R) >droSec1.super_2 5652389 93 - 7591821 ACUACAUCCACUUGACUCCAUG--GCGUUGUCGUCGCCAUCGUCGUCGACGUACCCGGCAAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........((((((((...(((--(((.......)))))).)))((((((......((((........)))).......))))))...))))).. ( -25.22, z-score = -1.90, R) >droYak2.chr3L 6283500 95 - 24197627 ACUACAUCCACUUGACUCCAUGGCGCGUUGUCGUCGGCAUCGUCGUCGACGUUCCCGGCAAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((......((.((((((.((.((....)).)).)))))).)).((((........))))..............))))))).. ( -26.70, z-score = -1.38, R) >dp4.chrXR_group6 5404481 86 + 13314419 ACUACAUCCACUUGACUCCA-----UGUCCUCGUCGUCCUUCUCCUCGUGACG----GCGAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((.....-----(((..(((((((((........).))))----))))..))).....((((....))))...))))))).. ( -23.30, z-score = -3.61, R) >droPer1.super_27 551143 86 - 1201981 ACUACAUCCACUUGACUCCA-----UGUCCUCGUCGUCCUUCUCCUCGUGACG----GCGAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((.....-----(((..(((((((((........).))))----))))..))).....((((....))))...))))))).. ( -23.30, z-score = -3.61, R) >droWil1.scaffold_180698 3976652 89 - 11422946 ACUACAUCCACUUGACUUCAGU--UCGAAAGAAAAGGCGGGGU--AUGAGGCAAU--GGAAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((..(((((--((....)))(((....(((--((...(((.(--....))))..)))))..)))...))))..))))))).. ( -23.70, z-score = -2.33, R) >droVir3.scaffold_13049 11670951 90 - 25233164 ACUACAAGCACUUGACUGCAGUUCGCUAGCUGCGCCUGUGCUCCAGC-----CCAGGACGCAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((..(((....((....((((((((.((....))-----.)))).)))).))...)))((((....))))...))))))).. ( -27.30, z-score = -1.43, R) >droMoj3.scaffold_6680 15609919 94 + 24764193 ACUGCAAGCACUUGACUGCAGCUCGCCCAGCUCUCCCA-GCUCCACCUGCUCCCACUAUGCAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ..((((.(((..((...((((.......((((.....)-)))....))))...))...))).)))).......(((((((.....)))))))... ( -18.40, z-score = -0.07, R) >droGri2.scaffold_15110 11130455 90 - 24565398 ACUACAAACACUUGACUGCUAGCUGCUCCCUUCACUCG-GCUGCCCCU----CCAGGGUGCAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((.(((.(((((...........))-)))(((((.----...))).))..))).....((((....))))...))))))).. ( -22.60, z-score = -1.28, R) >consensus ACUACAUCCACUUGACUCCAUG__GCGUUGUCGUCGUCAUCGUCGUCGACGCACCCGGCAAAUGCAAAUGCCAACUUAAGUUGAUUUUAAGUGAU ........(((((((................................................((....))((((....))))...))))))).. ( -9.70 = -9.70 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:44 2011