
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,703,142 – 5,703,234 |
| Length | 92 |
| Max. P | 0.688185 |

| Location | 5,703,142 – 5,703,234 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.48235 |
| G+C content | 0.55297 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -11.15 |
| Energy contribution | -10.82 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5703142 92 + 24543557 UGCUACUCCUGCCUUU-CAACCC-----------ACCGCCUCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAGCUCUCUGCAG .((.......))....-......-----------...((.((.....)).))........((((..(((((((((............)))))))).)..)))). ( -17.40, z-score = -0.59, R) >droPer1.super_27 536145 104 + 1201981 UGCUGCCUCUGCCCUCGCCCCCCCACGGGCCCCUGUCCGUGCCUUCUGCUGCUUUCAAUUUGCAAUGGCGGCUGUCAUUUCAGCUCAAUAGCAGCUCUCUCUCC .(((((.........((((....(((((((....)))))))........(((.........)))..))))((((......))))......)))))......... ( -31.60, z-score = -2.70, R) >dp4.chrXR_group6 5389781 103 - 13314419 UGCUGCCUCUGCCCUCGCCCCCC-ACGGACCCCUGUCCGUGCCUUCUGCUGCUUUCAAUUUGCAAUGGCGGCUGUCAUUUCAGCUCAAUAGCAGCUCUAUCUCC .(((((.........((((...(-((((((....)))))))........(((.........)))..))))((((......))))......)))))......... ( -32.20, z-score = -3.63, R) >droEre2.scaffold_4784 8393919 98 + 25762168 UGCUACUCCUGCUCUU-CAGCCCGGCC-----CCACCAACGCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAACUCACUGCCG (((((...........-.((((.(((.-----........)))......(((.........)))..))))((((......))))....)))))........... ( -19.30, z-score = -0.42, R) >droYak2.chr3L 6269352 103 + 24197627 UGCUACUCCCGCUUUU-CAGCCCAACCGCCCCCCACCGACGCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAGCUCACUGCAG .((.......(((...-.)))...............(((......)))..))........((((.((((((((((............)))))))).)).)))). ( -20.30, z-score = -0.61, R) >droSec1.super_2 5638946 79 + 7591821 UGCUACUCCUCCCUUU-CAACCC-----------ACCGACUCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAG------------- .((((...........-......-----------..(((......))).(((.........))).)))).((((......)))).......------------- ( -11.00, z-score = -1.10, R) >consensus UGCUACUCCUGCCCUU_CAACCC_AC______C_ACCGACGCCCCUCGAUGCUUUCAAUUUGCAAUGGCUGCUGUCAUUUCAGCUCAAUAGCAGCUCUCUGCCG (((((........................................(((.(((.........))).)))..((((......))))....)))))........... (-11.15 = -10.82 + -0.33)
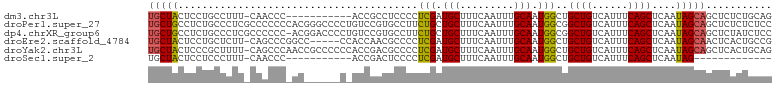

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:39 2011