| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,701,524 – 5,701,658 |
| Length | 134 |
| Max. P | 0.982134 |

| Location | 5,701,524 – 5,701,658 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 152 |
| Reading direction | forward |
| Mean pairwise identity | 70.98 |
| Shannon entropy | 0.52376 |
| G+C content | 0.36334 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.11 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
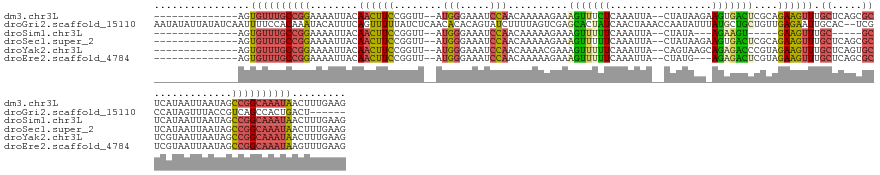
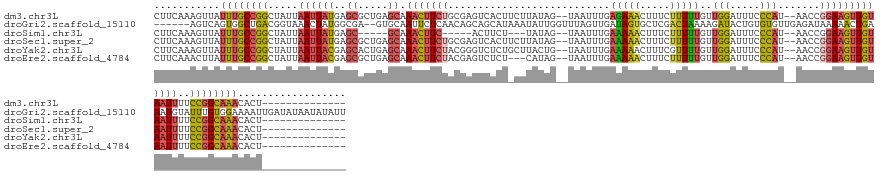
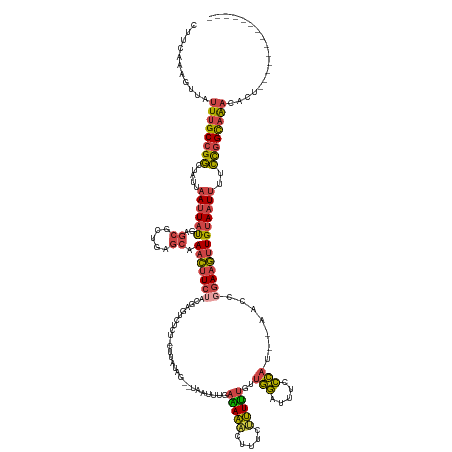
>dm3.chr3L 5701524 134 + 24543557 --------------AGUGUUUGCCGGAAAAUUACAACUUCCGGUU--AUGGGAAAUCCAACAAAAAGAAAGUUUCUCAAAUUA--CUAUAAGAAGUGACUCGCAGAAGUUUGCUCAGCGCUCAUAAUUAAUAGCCGGCAAAUAACUUUGAAG --------------..((((((((((((.........))))((((--(((((((((..............)))))))......--......((.(((.((.((((....))))..))))))).......))))))))))))))......... ( -30.84, z-score = -1.60, R) >droGri2.scaffold_15110 11114896 144 + 24565398 AAUAUAUUAUAUCAAUUUUCCACAAAUACAUUUCAGUUUUUAUCUCAACACACAGUAUCUUUUAGUCGAGCACUAUCAACUAAACCAAUAUUUAUGCUGCUGUUGAGAAUUGCAC--UCGCCAUAGUUUACCGUCAGCCACUGACU------ ..((.(((((................................((((((((..((((((..((((((.((......)).)))))).........)))))).))))))))...((..--..)).))))).))..(((((...))))).------ ( -23.10, z-score = -1.95, R) >droSim1.chr3L 5215940 121 + 22553184 --------------AGUGUUUGCCGGAAAAUUACAACUUCCGGUU--AUGGGAAAUCCAACAAAAAGAAAGUUUUUCAAAUUA--CUAUA---AGAAGU-----GAAGUUUGC-----GCUCAUAAUUAAUAGCCGGCAAAUAACUUUGAAG --------------..((((((((((((.........))))((((--((.((....))........((..((.(((((..((.--.....---.))..)-----))))...))-----..)).......))))))))))))))......... ( -25.40, z-score = -1.41, R) >droSec1.super_2 5637349 134 + 7591821 --------------AGUGUUUGCCGGAAAAUUACAACUUCCGGUU--AUGGGAAAUCCAACAAAAAGAAAGUUUUUCAAAUUA--CUAUAAGAAGUGACUCGCAGAAGUUUGCUCAGCGCUCAUAAUUAAUAGCCGGCAAAUAACUUUGAAG --------------..((((((((((((.........))))((((--(((..((((..............))))..)......--......((.(((.((.((((....))))..))))))).......))))))))))))))......... ( -28.34, z-score = -0.91, R) >droYak2.chr3L 6267680 134 + 24197627 --------------AGUGUUUGCCGGAAAAUUACAACUUCCGGUU--AUGGGAAAUCCAACAAAACGAAAGUUUUUCAAAUUA--CAGUAAGCAGAGACCCGUAGAAGUUUGCUCAGUGCUCGUAAUUAAUAGCCGGCAAAUAACUUUGAAG --------------..((((((((((..((((((((((((....(--(((((.........(((((....)))))........--..(....).....)))))))))))).((.....))..)))))).....))))))))))......... ( -35.60, z-score = -2.78, R) >droEre2.scaffold_4784 8392284 131 + 25762168 --------------AGUGUUUGCCGGAAAAUUACAACUUCCGGUU--AUGGGAAAUCCAACAAAAAGAAAGUUUUUCAAAUUA--CUAUG---AGAGACUCGUAGAAGUUUGCUCAGCGCUCGUAAUUAAUAGCCGGCAAAUAAGUUUGAAG --------------..((((((((((..((((((...(((((...--.))))).............((..(((...((((((.--(((((---((...))))))).))))))...)))..)))))))).....))))))))))......... ( -36.50, z-score = -3.26, R) >consensus ______________AGUGUUUGCCGGAAAAUUACAACUUCCGGUU__AUGGGAAAUCCAACAAAAAGAAAGUUUUUCAAAUUA__CUAUAAG_AGAGACUCGUAGAAGUUUGCUCAGCGCUCAUAAUUAAUAGCCGGCAAAUAACUUUGAAG ................((((((((((........((((((........(((.....)))..........(((((((.................)))))))....)))))).((.....)).............))))))))))......... (-15.05 = -15.11 + 0.06)
| Location | 5,701,524 – 5,701,658 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 152 |
| Reading direction | reverse |
| Mean pairwise identity | 70.98 |
| Shannon entropy | 0.52376 |
| G+C content | 0.36334 |
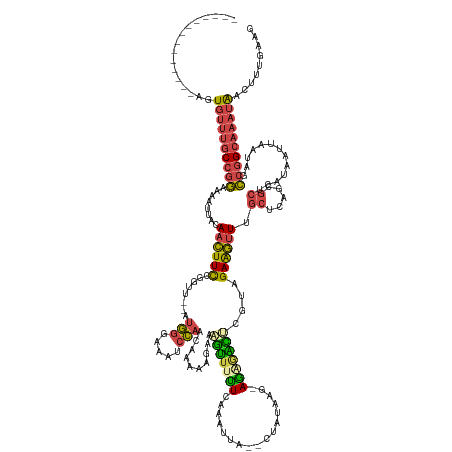
| Mean single sequence MFE | -31.51 |
| Consensus MFE | -16.34 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.982134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5701524 134 - 24543557 CUUCAAAGUUAUUUGCCGGCUAUUAAUUAUGAGCGCUGAGCAAACUUCUGCGAGUCACUUCUUAUAG--UAAUUUGAGAAACUUUCUUUUUGUUGGAUUUCCCAU--AACCGGAAGUUGUAAUUUUCCGGCAAACACU-------------- ...........((((((((.....((((((..((.....)).((((((((.((((...((((((...--.....)))))))))).....((((.((....)).))--)).))))))))))))))..))))))))....-------------- ( -31.50, z-score = -1.64, R) >droGri2.scaffold_15110 11114896 144 - 24565398 ------AGUCAGUGGCUGACGGUAAACUAUGGCGA--GUGCAAUUCUCAACAGCAGCAUAAAUAUUGGUUUAGUUGAUAGUGCUCGACUAAAAGAUACUGUGUGUUGAGAUAAAAACUGAAAUGUAUUUGUGGAAAAUUGAUAUAAUAUAUU ------.(((((...))))).........(.((((--(((((..((((((((((((......((((..(((((((((......))))))))).)))))))).))))))))............))))))))).)................... ( -34.24, z-score = -1.57, R) >droSim1.chr3L 5215940 121 - 22553184 CUUCAAAGUUAUUUGCCGGCUAUUAAUUAUGAGC-----GCAAACUUC-----ACUUCU---UAUAG--UAAUUUGAAAAACUUUCUUUUUGUUGGAUUUCCCAU--AACCGGAAGUUGUAAUUUUCCGGCAAACACU-------------- ...........((((((((..((((((((((((.-----.........-----....))---)))))--).........(((((((...((((.((....)).))--))..))))))).))))...))))))))....-------------- ( -24.84, z-score = -1.86, R) >droSec1.super_2 5637349 134 - 7591821 CUUCAAAGUUAUUUGCCGGCUAUUAAUUAUGAGCGCUGAGCAAACUUCUGCGAGUCACUUCUUAUAG--UAAUUUGAAAAACUUUCUUUUUGUUGGAUUUCCCAU--AACCGGAAGUUGUAAUUUUCCGGCAAACACU-------------- ...........((((((((.....((((((..((.....)).(((((((((((((.(((......))--).))))).............((((.((....)).))--)).))))))))))))))..))))))))....-------------- ( -31.00, z-score = -1.83, R) >droYak2.chr3L 6267680 134 - 24197627 CUUCAAAGUUAUUUGCCGGCUAUUAAUUACGAGCACUGAGCAAACUUCUACGGGUCUCUGCUUACUG--UAAUUUGAAAAACUUUCGUUUUGUUGGAUUUCCCAU--AACCGGAAGUUGUAAUUUUCCGGCAAACACU-------------- ...........((((((((.....((((((..(((.((((((....((....))....)))))).))--).........((((((((..((((.((....)).))--)).))))))))))))))..))))))))....-------------- ( -37.70, z-score = -3.61, R) >droEre2.scaffold_4784 8392284 131 - 25762168 CUUCAAACUUAUUUGCCGGCUAUUAAUUACGAGCGCUGAGCAAACUUCUACGAGUCUCU---CAUAG--UAAUUUGAAAAACUUUCUUUUUGUUGGAUUUCCCAU--AACCGGAAGUUGUAAUUUUCCGGCAAACACU-------------- ...........((((((((.....((((((..((..((((...((((....))))..))---))..)--).........(((((((...((((.((....)).))--))..)))))))))))))..))))))))....-------------- ( -29.80, z-score = -2.27, R) >consensus CUUCAAAGUUAUUUGCCGGCUAUUAAUUAUGAGCGCUGAGCAAACUUCUACGAGUCUCU_CUUAUAG__UAAUUUGAAAAACUUUCUUUUUGUUGGAUUUCCCAU__AACCGGAAGUUGUAAUUUUCCGGCAAACACU______________ ...........((((((((.....((((((..((.....))......................................(((((((.......(((.....))).......)))))))))))))..)))))))).................. (-16.34 = -17.07 + 0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:39 2011