
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,613,581 – 5,613,690 |
| Length | 109 |
| Max. P | 0.884700 |

| Location | 5,613,581 – 5,613,690 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.52692 |
| G+C content | 0.39032 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -11.43 |
| Energy contribution | -10.97 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
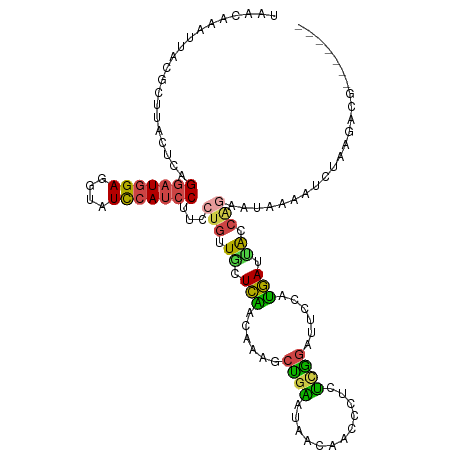
>dm3.chr3L 5613581 109 + 24543557 UAACAAAUUACGCGUACACAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUCAACAAAGCUGAAUAACAACCCUCUCGGAUUCCAUGAUUACCAAAAUAAAAUCUAAGACG------- ...........(((.(((.((((((((....))))))))...)))))).......................(((..((((..((..........))..))))..)))..------- ( -20.00, z-score = -1.40, R) >droSim1.chr3L 5131852 109 + 22553184 AAACAAAUUACGCUUACUCAGGAUGGAGGUAUCCAUCCUUCCUGUUCCUCGACAAAGCUGAAUAACAACCCUCUCGGAUUCCAUGAUUACCGGAAUAAAAUCUAAGACG------- ...........((((....((((((((....))))))))...((((....)))))))).............(((..((((((.........))).....)))..)))..------- ( -21.40, z-score = -1.00, R) >droSec1.super_2 5550080 109 + 7591821 AAACAAAUUACGCUUAUUCAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUCAACAAAGCUGAAUAACAACCCUCUCGGAUUCCAUGAUUACCAGAAUAAAAUCUAAGACG------- ...........(.((((((((((((((....)))))))........(((......))))))))))).....(((..((((...((.....))......))))..)))..------- ( -21.80, z-score = -1.72, R) >droYak2.chr3L 6178098 109 + 24197627 UAACAAAUUACGCUUACUCAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUCAACAAAGCCGAAUAGCAACCCUCUCGGAUUCCCUGAUUGCCAGAGUAAAAUCUAUGACC------- .............((((((((((((((....))))))))....((((((...(......)...))))))....((((.....))))......))))))...........------- ( -25.20, z-score = -1.64, R) >droEre2.scaffold_4784 8301359 109 + 25762168 UAACAAAUUACGUUUGCUCAGGAUGGAGGUAUCCAUCCAUCCUGUUGCUCAACAAAGCUGGAUAAUAAUCCUCUUGGAUUCCCUGAUCACCAGAAUAAAAUCUAAGACC------- ...........(((.((.(((((((((((...)).)))))))))..))..)))......((((....))))(((((((((..(((.....))).....)))))))))..------- ( -31.10, z-score = -3.40, R) >droWil1.scaffold_180698 7581372 108 - 11422946 --------CAAACUGACAAUGAAUUGAGGAGUUUAAGCUUCCACAUUUUUUAAAAUAAUGUGUGGAAUUCUUAAGAAACAACAAAAGGAUUCACAUUUAAAGUAAAUCUACAUAUA --------..........(((..(((..(..((((((.((((((((.((.......)).))))))))..))))))...)..)))..(((((.((.......)).))))).)))... ( -16.80, z-score = -0.75, R) >consensus UAACAAAUUACGCUUACUCAGGAUGGAGGUAUCCAUCCUUCCUGUUGCUCAACAAAGCUGAAUAACAACCCUCUCGGAUUCCAUGAUUACCAGAAUAAAAUCUAAGACG_______ ....................(((((((....)))))))...(((.((.(((......((((............))))......))).)).)))....................... (-11.43 = -10.97 + -0.46)


| Location | 5,613,581 – 5,613,690 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.52692 |
| G+C content | 0.39032 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -15.64 |
| Energy contribution | -14.45 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
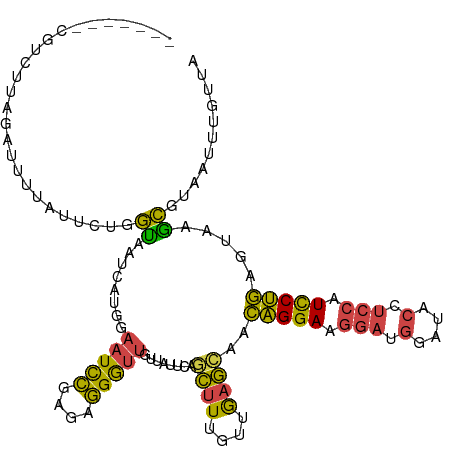
>dm3.chr3L 5613581 109 - 24543557 -------CGUCUUAGAUUUUAUUUUGGUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGUGUACGCGUAAUUUGUUA -------(.(((..((((((((..(....)..))))))))..))).)(((((...(((((...))))))))))....((((((((....))))))))................... ( -29.10, z-score = -1.43, R) >droSim1.chr3L 5131852 109 - 22553184 -------CGUCUUAGAUUUUAUUCCGGUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUCGAGGAACAGGAAGGAUGGAUACCUCCAUCCUGAGUAAGCGUAAUUUGUUU -------((.((((......((((((.......)))))).(((.(((((((.....))))))).))).....(((((.(((.((...))))).)))))..)))))).......... ( -29.20, z-score = -0.91, R) >droSec1.super_2 5550080 109 - 7591821 -------CGUCUUAGAUUUUAUUCUGGUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGAAUAAGCGUAAUUUGUUU -------((.((((.......(((((..........(((((....)))))........((((....))))..)))))((((((((....))))))))...)))))).......... ( -29.80, z-score = -1.63, R) >droYak2.chr3L 6178098 109 - 24197627 -------GGUCAUAGAUUUUACUCUGGCAAUCAGGGAAUCCGAGAGGGUUGCUAUUCGGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGAGUAAGCGUAAUUUGUUA -------....(((((((...(((((.....)))))(((((....)))))(((((((.((((....)))).......((((((((....)))))))))))).)))..))))))).. ( -34.10, z-score = -1.69, R) >droEre2.scaffold_4784 8301359 109 - 25762168 -------GGUCUUAGAUUUUAUUCUGGUGAUCAGGGAAUCCAAGAGGAUUAUUAUCCAGCUUUGUUGAGCAACAGGAUGGAUGGAUACCUCCAUCCUGAGCAAACGUAAUUUGUUA -------(((((((((......))))).)))).((((((((....)))))....))).((((....))))..(((((((((.((...)))))))))))((((((.....)))))). ( -35.00, z-score = -2.62, R) >droWil1.scaffold_180698 7581372 108 + 11422946 UAUAUGUAGAUUUACUUUAAAUGUGAAUCCUUUUGUUGUUUCUUAAGAAUUCCACACAUUAUUUUAAAAAAUGUGGAAGCUUAAACUCCUCAAUUCAUUGUCAGUUUG-------- ........(((((((.......))))))).....((((....(((((..(((((((...............))))))).)))))......))))..............-------- ( -15.56, z-score = -0.59, R) >consensus _______CGUCUUAGAUUUUAUUCUGGUAAUCAUGGAAUCCGAGAGGGUUGUUAUUCAGCUUUGUUGAGCAACAGGAAGGAUGGAUACCUCCAUCCUGAGUAAGCGUAAUUUGUUA ..........................((........(((((....)))))........((((....))))..(((((.(((.(....).))).))))).....))........... (-15.64 = -14.45 + -1.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:34 2011