| Sequence ID | dm3.chr3L |
|---|---|
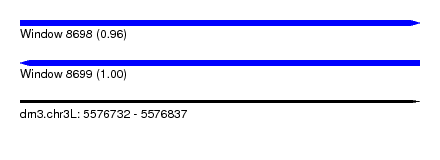
| Location | 5,576,732 – 5,576,837 |
| Length | 105 |
| Max. P | 0.997531 |

| Location | 5,576,732 – 5,576,837 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Shannon entropy | 0.48471 |
| G+C content | 0.45449 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -15.83 |
| Energy contribution | -17.17 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
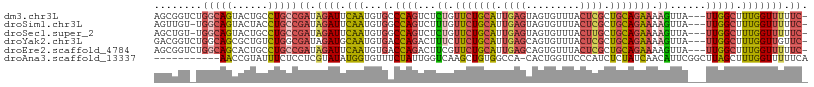
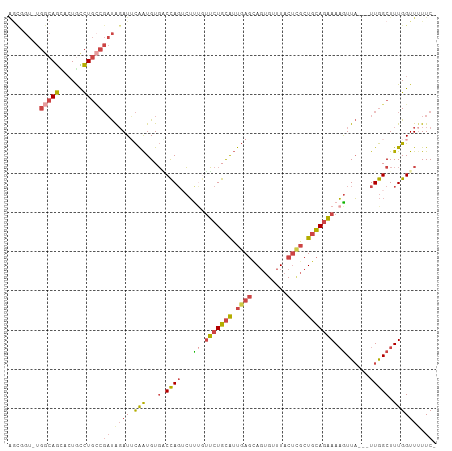
>dm3.chr3L 5576732 105 + 24543557 AGCGGUCUGGCAGUACUGCCUGCCGAUAGAUUCAAUGUGCCCAGUCUCUGUUCUGCAUUGAGUAGUGUUUACUCGCUGCAGAAAAGUUA---UUGGCUUUGGUUUUUC- ..((((..((((....)))).))))......((((...(((.(((..((.(((((((.(((((((...))))))).))))))).))..)---))))).))))......- ( -36.30, z-score = -2.92, R) >droSim1.chr3L 5092399 104 + 22553184 AGUUGU-UGGCAGUACUACCUGCCGAUAGAUUCAAUGUGGCCAGUCUUUGUUCUGCAUUGAGUAGUGUUUACUCGCUGCAGAAAAGUUA---UUGGCUUUGGUUUUUC- ..((((-((((((......))))))))))..((((...(((((((..((.(((((((.(((((((...))))))).))))))).))..)---))))))))))......- ( -38.80, z-score = -4.69, R) >droSec1.super_2 5513136 104 + 7591821 AGCUGU-UGGCAGUACUGCCUGCCGAUAGAUUCAAUGUGGCCAGUCUCUGUUCUGCAUUGAGUAGUGUUUACUUGCUGCAGAAAAGUUA---UUGGCUUUGGUUUUUC- ..((((-((((((......))))))))))..((((...(((((((..((.(((((((..((((((...))))))..))))))).))..)---))))))))))......- ( -40.50, z-score = -4.39, R) >droYak2.chr3L 6141578 105 + 24197627 GACGGUCUGGCAGCGCUGUCUGGCGAUAGAUGCAAUGUGACCAGACUUUCUUCUGCAUUGAGCAGUGUUUACUCGCUGCAGAAAAGUUA---UUGGCUUUGGUUGUUC- ((.(((((((((((((((((....))))).)))....)).))))))).)).((((((.((((.........)))).))))))(((((..---...)))))........- ( -33.30, z-score = -0.78, R) >droEre2.scaffold_4784 8263448 105 + 25762168 AGCGGUCUGGCAGCACUGCCUGCCGAUAGAUUCAAUGUGACCAGACUUCGUUCUGCAUUGAGCAGUGUUUACUCGCUGCAGAAAAGUUA---UUGGCUUUGGUUUUUC- (((((((.(((((......)))))((.....)).....)))).(((((..(((((((.((((.........)))).)))))))))))).---...)))..........- ( -31.60, z-score = -0.74, R) >droAna3.scaffold_13337 10320292 97 - 23293914 -----------AACCGUAUUUCUCCUCGUAUAUGGUGUUUCUAUUGGUCAAGCUGUGGCCA-CACUGGUUCCCAUCUCUAUCAACAUUCGGCUUAGCUUUGGUUUUUCA -----------.(((((((..........)))))))....(((..((..((((((((....-)))(((...)))...............)))))..)).)))....... ( -16.30, z-score = 0.05, R) >consensus AGCGGU_UGGCAGCACUGCCUGCCGAUAGAUUCAAUGUGACCAGUCUUUGUUCUGCAUUGAGCAGUGUUUACUCGCUGCAGAAAAGUUA___UUGGCUUUGGUUUUUC_ ........(((((......)))))..............((((.........((((((.((((.........)))).))))))(((((((....)))))))))))..... (-15.83 = -17.17 + 1.34)
| Location | 5,576,732 – 5,576,837 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Shannon entropy | 0.48471 |
| G+C content | 0.45449 |
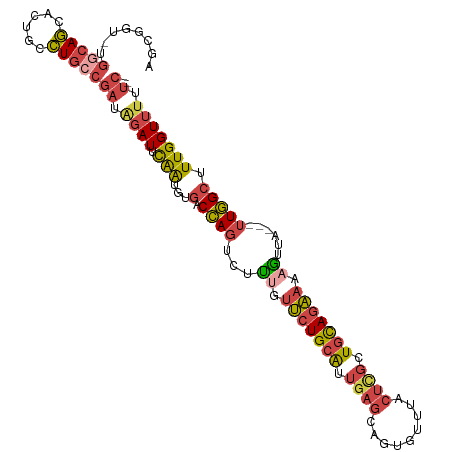
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5576732 105 - 24543557 -GAAAAACCAAAGCCAA---UAACUUUUCUGCAGCGAGUAAACACUACUCAAUGCAGAACAGAGACUGGGCACAUUGAAUCUAUCGGCAGGCAGUACUGCCAGACCGCU -...........(((..---...((.(((((((..(((((.....)))))..))))))).))......)))..............(((((......)))))........ ( -31.70, z-score = -3.47, R) >droSim1.chr3L 5092399 104 - 22553184 -GAAAAACCAAAGCCAA---UAACUUUUCUGCAGCGAGUAAACACUACUCAAUGCAGAACAAAGACUGGCCACAUUGAAUCUAUCGGCAGGUAGUACUGCCA-ACAACU -...........((((.---......(((((((..(((((.....)))))..))))))).......))))...............(((((......))))).-...... ( -30.24, z-score = -4.60, R) >droSec1.super_2 5513136 104 - 7591821 -GAAAAACCAAAGCCAA---UAACUUUUCUGCAGCAAGUAAACACUACUCAAUGCAGAACAGAGACUGGCCACAUUGAAUCUAUCGGCAGGCAGUACUGCCA-ACAGCU -...........((((.---(..((.(((((((...((((.....))))...))))))).))..).))))...............(((((......))))).-...... ( -28.50, z-score = -3.14, R) >droYak2.chr3L 6141578 105 - 24197627 -GAACAACCAAAGCCAA---UAACUUUUCUGCAGCGAGUAAACACUGCUCAAUGCAGAAGAAAGUCUGGUCACAUUGCAUCUAUCGCCAGACAGCGCUGCCAGACCGUC -..........((((..---....(((((((((..(((((.....)))))..)))))))))..(((((((...............))))))).).)))........... ( -29.56, z-score = -2.42, R) >droEre2.scaffold_4784 8263448 105 - 25762168 -GAAAAACCAAAGCCAA---UAACUUUUCUGCAGCGAGUAAACACUGCUCAAUGCAGAACGAAGUCUGGUCACAUUGAAUCUAUCGGCAGGCAGUGCUGCCAGACCGCU -..........(((...---..(((((((((((..(((((.....)))))..))))))..)))))..((((.....((.....))(((((......))))).))))))) ( -32.90, z-score = -2.67, R) >droAna3.scaffold_13337 10320292 97 + 23293914 UGAAAAACCAAAGCUAAGCCGAAUGUUGAUAGAGAUGGGAACCAGUG-UGGCCACAGCUUGACCAAUAGAAACACCAUAUACGAGGAGAAAUACGGUU----------- ................(((((..(((((...(((.((((..((....-.)))).)).)))...)))))......((........)).......)))))----------- ( -14.70, z-score = 0.14, R) >consensus _GAAAAACCAAAGCCAA___UAACUUUUCUGCAGCGAGUAAACACUACUCAAUGCAGAACAAAGACUGGCCACAUUGAAUCUAUCGGCAGGCAGUACUGCCA_ACCGCU ............((((.......((((((((((..(((((.....)))))..))))))..))))..))))...............(((((......)))))........ (-16.79 = -16.93 + 0.15)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:31 2011