
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,536,590 – 5,536,700 |
| Length | 110 |
| Max. P | 0.999734 |

| Location | 5,536,590 – 5,536,700 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Shannon entropy | 0.35924 |
| G+C content | 0.48338 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -15.75 |
| Energy contribution | -17.71 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
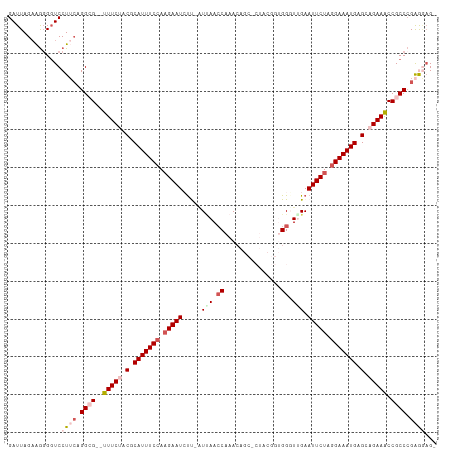
>dm3.chr3L 5536590 110 + 24543557 GAUUAGAAGGGGUCCUUGAGGCG--UUUCUACGCAUUUCCAAGAAUCUUCAUUAAACAAACAGCCCUACGGUGGGUUGAAUUCUAAGAAAUGAGCAGAAACCGCCCGAGGAG- ............((((((.((((--(((((.(.((((((..(((((..............((((((......)))))).)))))..)))))).).))))).)))))))))).- ( -37.06, z-score = -3.24, R) >droEre2.scaffold_4784 8223094 98 + 25762168 GAUUAGAGGCGGUCCUACUGGCG--UUUCCACGCAUUUCCACGAAUGUC-AUCAACCAAAC-----------GGUUGGAAUUCUAGGAAAUGCGCAGAAACCGCCCGAAAAG- .....(.(((((.((....)))(--((((..(((((((((..((((...-.((((((....-----------)))))).))))..)))))))))..))))))))))......- ( -36.30, z-score = -4.22, R) >droYak2.chr3L 6099865 104 + 24197627 GAUUAGCAAGGCUCCUACUGGCGCAUUUCUACGCAUUUCCAAGAAUGCCAUUAAACCCAAACG---------GGGUUGAAUUCUAGGAAAUGAGCAGAAACCGCCAGGAUCGG ((((....((....)).((((((..(((((.(.(((((((.(((((.......(((((.....---------)))))..))))).))))))).).))))).)))))))))).. ( -37.30, z-score = -3.99, R) >droSec1.super_2 5473168 109 + 7591821 GAUUAGAAGGGGUCCUUCAGGGG--UUUCUACGCAUUUCCAAGAAUCUU-GUUAACCAAACAGCUCUACGGUGGGUGGAAUUCUAGGAAAUGAGCAGAAGCCGCCCGAGGAG- ............(((((..((((--(((((.(.(((((((.(((((.(.-.(..(((............)))..)..).))))).))))))).).))))))).)).))))).- ( -36.70, z-score = -1.77, R) >droSim1.chr3L 5051789 109 + 22553184 GAUUAGAAGGGGUCCUUCAGGGG--UUUCCACGCAUUUCCAAGAAUCUU-GUUAACCAAACAGCACUACGGUGGUUUGAAUUCUAGGAAAUGAGCAGAAACCGCCCGAGGAG- ............(((((..((((--((((..(.(((((((.(((((.((-(((.....)))))(((....)))......))))).))))))).)..)))))).)).))))).- ( -34.90, z-score = -2.18, R) >consensus GAUUAGAAGGGGUCCUUCAGGCG__UUUCUACGCAUUUCCAAGAAUCUU_AUUAACCAAACAGC_CUACGGUGGGUUGAAUUCUAGGAAAUGAGCAGAAACCGCCCGAGGAG_ ..............((((.((((...((((.(.(((((((.(((((.................................))))).))))))).).))))..)))).))))... (-15.75 = -17.71 + 1.96)


| Location | 5,536,590 – 5,536,700 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Shannon entropy | 0.35924 |
| G+C content | 0.48338 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -25.02 |
| Energy contribution | -26.02 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.60 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
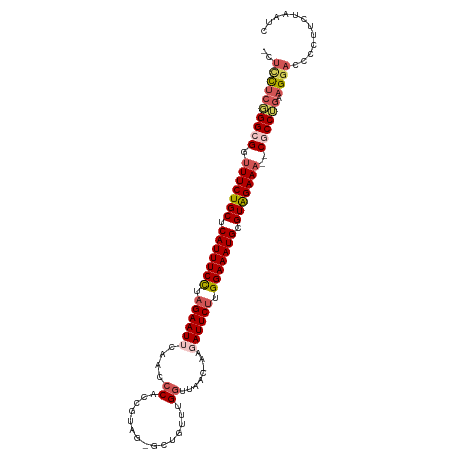
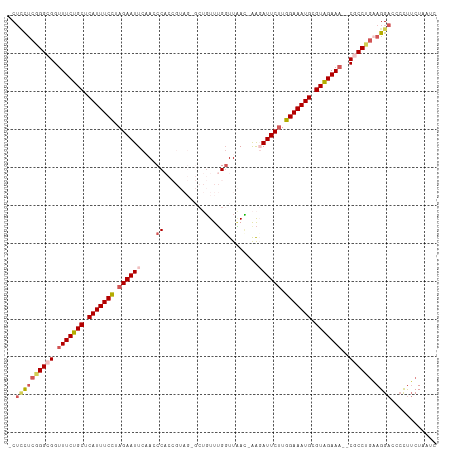
>dm3.chr3L 5536590 110 - 24543557 -CUCCUCGGGCGGUUUCUGCUCAUUUCUUAGAAUUCAACCCACCGUAGGGCUGUUUGUUUAAUGAAGAUUCUUGGAAAUGCGUAGAAA--CGCCUCAAGGACCCCUUCUAAUC -.((((.(((((.(((((((.(((((((.((((((((.(((......))).)).............)))))).))))))).)))))))--)))).).))))............ ( -37.21, z-score = -3.63, R) >droEre2.scaffold_4784 8223094 98 - 25762168 -CUUUUCGGGCGGUUUCUGCGCAUUUCCUAGAAUUCCAACC-----------GUUUGGUUGAU-GACAUUCGUGGAAAUGCGUGGAAA--CGCCAGUAGGACCGCCUCUAAUC -......((((((((.((((((((((((..(((((((((((-----------....)))))..-)).))))..))))))))(((....--)))..))))))))))))...... ( -43.20, z-score = -5.41, R) >droYak2.chr3L 6099865 104 - 24197627 CCGAUCCUGGCGGUUUCUGCUCAUUUCCUAGAAUUCAACCC---------CGUUUGGGUUUAAUGGCAUUCUUGGAAAUGCGUAGAAAUGCGCCAGUAGGAGCCUUGCUAAUC ((....((((((((((((((.(((((((.((((((((((((---------.....))))....))).))))).))))))).)))))))).))))))..))(((...))).... ( -41.70, z-score = -4.58, R) >droSec1.super_2 5473168 109 - 7591821 -CUCCUCGGGCGGCUUCUGCUCAUUUCCUAGAAUUCCACCCACCGUAGAGCUGUUUGGUUAAC-AAGAUUCUUGGAAAUGCGUAGAAA--CCCCUGAAGGACCCCUUCUAAUC -.((((((((.((.((((((.(((((((.((((((......((((..........))))....-..)))))).))))))).)))))).--)))))).))))............ ( -38.70, z-score = -4.17, R) >droSim1.chr3L 5051789 109 - 22553184 -CUCCUCGGGCGGUUUCUGCUCAUUUCCUAGAAUUCAAACCACCGUAGUGCUGUUUGGUUAAC-AAGAUUCUUGGAAAUGCGUGGAAA--CCCCUGAAGGACCCCUUCUAAUC -.((((((((.((((((..(.(((((((.((((((((((((((....)))..)))))......-..)))))).))))))).)..))))--)))))).))))............ ( -42.10, z-score = -5.22, R) >consensus _CUCCUCGGGCGGUUUCUGCUCAUUUCCUAGAAUUCAACCCACCGUAG_GCUGUUUGGUUAAC_AAGAUUCUUGGAAAUGCGUAGAAA__CGCCUGAAGGACCCCUUCUAAUC ..((((((((((..((((((.(((((((.((((((...............................)))))).))))))).))))))...)))))).))))............ (-25.02 = -26.02 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:27 2011