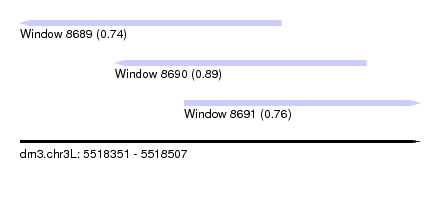
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,518,351 – 5,518,507 |
| Length | 156 |
| Max. P | 0.889364 |

| Location | 5,518,351 – 5,518,453 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.42 |
| Shannon entropy | 0.49968 |
| G+C content | 0.42399 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
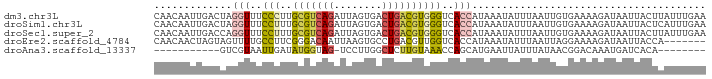
>dm3.chr3L 5518351 102 - 24543557 UCACUAAUCUGACGCAAGGGAAACCUAGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAAU----------AAAAAACACCCAUCUGGUCUAGACAGCUCUAGGCCC .........((((...(((....))).)))).(((((((((.(..........).)))))))))..----------...............((((((((....)))))))). ( -30.90, z-score = -3.99, R) >droSim1.chr3L 5034012 102 - 22553184 UCACUAAUCUGACGCAAAGGAAACCUAGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAAU----------AAAAAACACCCAUCUGGUCUAGACAGUUCCAGGCCC .........((((.....(....)...)))).(((((((((.(..........).)))))))))..----------...............(((((.((....)).))))). ( -21.10, z-score = -1.23, R) >droSec1.super_2 5454626 102 - 7591821 UCACUAAUCUGACGCAAAGGAAACCUGGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAAU----------AAAAAACACCCAUCUGGUCUAGACAGUUCCAGGCCC .........((((.((..(....).)))))).(((((((((.(..........).)))))))))..----------...............(((((.((....)).))))). ( -21.80, z-score = -0.86, R) >droYak2.chr3L 6081474 94 - 24197627 ------------------GCGAAACUACUAAUUUGUUGAUGUCUUUAGCAACUGUCAUCAACGACUUGAUGUGCUGAACAGACAUUGAUCAGAUCUAGACACUUUCAGGCUC ------------------..((((...(((((((((..((((((((((((.....((((((....))))))))))))..))))))..).))))).)))....))))...... ( -24.40, z-score = -1.86, R) >droEre2.scaffold_4784 8202723 102 - 25762168 GCACUUAAUUGUCCCGAAGGCAAAACUACUAGUUGUUGAUGUCUUCAACGACUUGAUUUGCUCAGU----------AAAAAACACCCAUCAGGUCUAGACAGAUCCAGGCUC ((......(((((..(((((((.(((........)))..)))))))...((((((((((((...))----------)).........))))))))..)))))......)).. ( -23.80, z-score = -1.32, R) >consensus UCACUAAUCUGACGCAAAGGAAACCUAGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAAU__________AAAAAACACCCAUCUGGUCUAGACAGUUCCAGGCCC ................................(((((((((.(..........).)))))))))...........................(((((.((....)).))))). (-10.48 = -10.80 + 0.32)



| Location | 5,518,388 – 5,518,486 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 65.05 |
| Shannon entropy | 0.62210 |
| G+C content | 0.38446 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5518388 98 - 24543557 UCACAAUUAAAUAUUUAUGGUGACCCACGUCAGUCACUAA--UCUGACGCAAGGGAAACCUAGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAA .................(((((((........))))))).--..((((...(((....))).)))).(((((((((.(..........).))))))))). ( -28.30, z-score = -3.55, R) >droSim1.chr3L 5034049 98 - 22553184 UCACAAUUAAAUAUUUAUGGUGACCCACGUCAGUCACUAA--UCUGACGCAAAGGAAACCUAGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAA ..................(((..((..((((((.......--.))))))....))..))).......(((((((((.(..........).))))))))). ( -23.30, z-score = -2.28, R) >droSec1.super_2 5454663 98 - 7591821 UCACAAUUAAAUAUUUAUGGUGACCCACGUCAGUCACUAA--UCUGACGCAAAGGAAACCUGGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAA ....................(((((..((((((.......--.))))))....(....)..))))).(((((((((.(..........).))))))))). ( -25.80, z-score = -2.64, R) >droEre2.scaffold_4784 8202762 96 - 25762168 UCCUAAUUAAAUAUUUAUGGUGACCAACGUCAGGCACUUA--AUUGUCCCGAAGGCAAAACUACUAGUUGUUGAUGUCUUCAACGACU-UGAUUUGCUC- .................(((((((....))))((((....--..)))))))..((((((.(....(((((((((.....)))))))))-.).)))))).- ( -21.70, z-score = -1.81, R) >droAna3.scaffold_13337 10257876 99 + 23293914 CCGUUAUAAAUAAUUCAUGCUGGUUUACAAGAGCCAAGGACUACCAUAUCAAUUACGACGUAUAUCUUCAACGACUUUCAUUAGGGACAUGACUUGCUG- ..(((((.........((((((((((....)))))).((....))..............)))).(((..((.(.....).))..))).)))))......- ( -11.90, z-score = 1.19, R) >consensus UCACAAUUAAAUAUUUAUGGUGACCCACGUCAGUCACUAA__UCUGACGCAAAGGAAACCUAGUCAAUUGUUGAUGUCUUUAGCAACUGUCAUCAACGAA .................(((((((........)))))))............................(((((((.....))))))).............. ( -8.52 = -8.76 + 0.24)


| Location | 5,518,415 – 5,518,507 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 65.17 |
| Shannon entropy | 0.63928 |
| G+C content | 0.34498 |
| Mean single sequence MFE | -18.56 |
| Consensus MFE | -7.54 |
| Energy contribution | -8.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5518415 92 + 24543557 CAACAAUUGACUAGGUUUCCCUUGCGUCAGAUUAGUGACUGACGUGGGUCACCAUAAAUAUUUAAUUGUGAAAAGAUAAUUACUUAUUUGAA .............(((..(((..(((((((........))))))))))..))).((((((..(((((((......)))))))..)))))).. ( -25.10, z-score = -3.38, R) >droSim1.chr3L 5034076 92 + 22553184 CAACAAUUGACUAGGUUUCCUUUGCGUCAGAUUAGUGACUGACGUGGGUCACCAUAAAUAUUUAAUUGUGAAAAGAUAAUUACUCAUUUGAA ...(((.(((...(((..(((..(((((((........))))))))))..))).........(((((((......))))))).))).))).. ( -22.60, z-score = -2.30, R) >droSec1.super_2 5454690 92 + 7591821 CAACAAUUGACCAGGUUUCCUUUGCGUCAGAUUAGUGACUGACGUGGGUCACCAUAAAUAUUUAAUUGUGAAAAGAUAAUUACUUAUUUGAA .............(((..(((..(((((((........))))))))))..))).((((((..(((((((......)))))))..)))))).. ( -23.40, z-score = -2.67, R) >droEre2.scaffold_4784 8202787 85 + 25762168 CAACAACUAGUAGUUUUGCCUUCGGGACAAUUAAGUGCCUGACGUUGGUCACCAUAAAUAUUUAAUUAGGAAAAGAUAAUUACCA------- ((((..(..(((.(((((((....)).)))..)).)))..)..))))...............((((((........))))))...------- ( -12.90, z-score = 0.39, R) >droAna3.scaffold_13337 10257916 72 - 23293914 -----------GUCGUAAUUGAUAUGGUAG-UCCUUGGCUCUUGUAAACCAGCAUGAAUUAUUUAUAACGGACAAAUGAUCACA-------- -----------(((((((((.((.((((((-(.....))).......))))..)).))))))........)))...........-------- ( -8.81, z-score = 1.27, R) >consensus CAACAAUUGACUAGGUUUCCUUUGCGUCAGAUUAGUGACUGACGUGGGUCACCAUAAAUAUUUAAUUGUGAAAAGAUAAUUACUUAUUUGAA .............(((..(((..(((((((........))))))))))..)))....................................... ( -7.54 = -8.14 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:24 2011