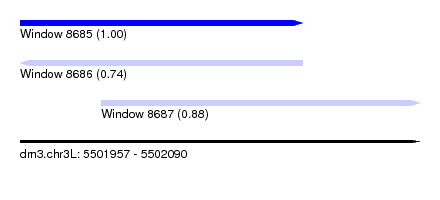
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,501,957 – 5,502,090 |
| Length | 133 |
| Max. P | 0.995964 |

| Location | 5,501,957 – 5,502,051 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 68.46 |
| Shannon entropy | 0.60170 |
| G+C content | 0.57516 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5501957 94 + 24543557 UGCAAGUCAGCACACAAAAUGCAGCAGAACGAGCGGUGGCAGAUGGAA---UUGGUCCUGGAAACAGGACGCUGUGGCGGGA---AUCCUGCAGUGCCAC .........((((.......(((((.......((....))........---...((((((....))))))))))).(((((.---..))))).))))... ( -35.00, z-score = -2.07, R) >dp4.chrXR_group6 5161415 99 - 13314419 UCAAAGUCAGCAACACUCAACAUCCACCAGAUGCAGAUGCGGAUGCGGAUGCGGAUGC-AGAAGGAGCGCCUGCUGGCGGGAUGGGAACUGCAGUGCGAC .....(((.(((.((.....(((((.((...((((..((((........))))..)))-)...))..((((....))))))))).....))...)))))) ( -31.70, z-score = -0.19, R) >droSim1.chr3L 5017633 94 + 22553184 UGCAAGUCAGCACACAGAAUGCAGCAGAAUGAGCGGUGGCAGAUGGAA---GCGGUCCUGGAAACAGGACGCUGAGGCGGGA---AUCCUGCAGUGCCAC .(((..(((((...((...(((.((.......))....)))..))...---...((((((....))))))))))).(((((.---..)))))..)))... ( -35.70, z-score = -2.30, R) >droSec1.super_2 5438357 94 + 7591821 UGCAAGUCAGCACACAGAAUGCAGCAGAAUGAGCGGUGGCAGAUGGAA---GCGGUCCUGGAAACAGGACGCUGAGGCGGGA---AUCCUGCAGUACCAC (((...(((((...((...(((.((.......))....)))..))...---...((((((....))))))))))).(((((.---..))))).))).... ( -34.30, z-score = -2.41, R) >droYak2.chr3L 6064384 97 + 24197627 UCCAAGUCAGCACAACAAAUGCAGCAGGAUGAGCGGUGGCAGAUGCGGUGGCAGGUCCUUGCAACAGGACGAUGAGGCGGGA---AUCCUGCAGUGCCAC .........((((......((((..(((((..((.((.((....)).)).))..))))))))).............(((((.---..))))).))))... ( -28.10, z-score = 1.02, R) >droEre2.scaffold_4784 8186773 74 + 25762168 -------------------UCCAA-AAUGCAUAUGGUGGCAUAUGGAG---CAGGUCCUGGAAACAGGACGCUGAGGCGGGA---AUCCUGCAGUGCCAC -------------------.....-..........(((((((......---(((((((((....)))))).)))..(((((.---..))))).))))))) ( -33.50, z-score = -4.03, R) >consensus UGCAAGUCAGCACACAAAAUGCAGCAGAAUGAGCGGUGGCAGAUGGAA___CAGGUCCUGGAAACAGGACGCUGAGGCGGGA___AUCCUGCAGUGCCAC ...................................((((((.............((((((....))))))......(((((......)))))..)))))) (-24.16 = -24.50 + 0.34)
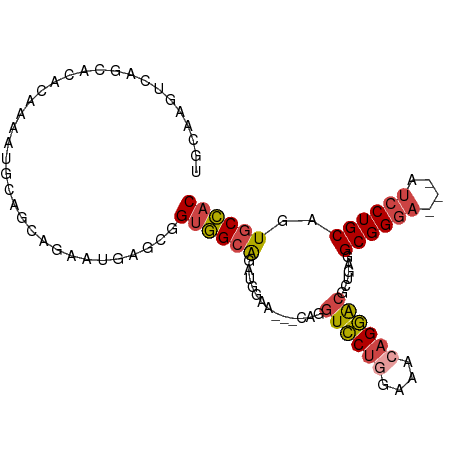
| Location | 5,501,957 – 5,502,051 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 68.46 |
| Shannon entropy | 0.60170 |
| G+C content | 0.57516 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -15.95 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5501957 94 - 24543557 GUGGCACUGCAGGAU---UCCCGCCACAGCGUCCUGUUUCCAGGACCAA---UUCCAUCUGCCACCGCUCGUUCUGCUGCAUUUUGUGUGCUGACUUGCA ((((((..((((((.---...(((....))))))))).....(((....---.)))...)))))).((..(((..((.((((...)))))).)))..)). ( -28.00, z-score = -0.84, R) >dp4.chrXR_group6 5161415 99 + 13314419 GUCGCACUGCAGUUCCCAUCCCGCCAGCAGGCGCUCCUUCU-GCAUCCGCAUCCGCAUCCGCAUCUGCAUCUGGUGGAUGUUGAGUGUUGCUGACUUUGA ((.(((((((((.........((((....))))......))-)))((.((((((((....((....)).....)))))))).)))))).))......... ( -31.76, z-score = -0.46, R) >droSim1.chr3L 5017633 94 - 22553184 GUGGCACUGCAGGAU---UCCCGCCUCAGCGUCCUGUUUCCAGGACCGC---UUCCAUCUGCCACCGCUCAUUCUGCUGCAUUCUGUGUGCUGACUUGCA ((((((..((.((..---..))))...(((((((((....))))).)))---)......)))))).((....((.((.((((...)))))).))...)). ( -28.40, z-score = -0.86, R) >droSec1.super_2 5438357 94 - 7591821 GUGGUACUGCAGGAU---UCCCGCCUCAGCGUCCUGUUUCCAGGACCGC---UUCCAUCUGCCACCGCUCAUUCUGCUGCAUUCUGUGUGCUGACUUGCA ((((((..((.((..---..))))...(((((((((....))))).)))---)......)))))).((....((.((.((((...)))))).))...)). ( -26.40, z-score = -0.53, R) >droYak2.chr3L 6064384 97 - 24197627 GUGGCACUGCAGGAU---UCCCGCCUCAUCGUCCUGUUGCAAGGACCUGCCACCGCAUCUGCCACCGCUCAUCCUGCUGCAUUUGUUGUGCUGACUUGGA ((((((.(((.((..---..))........(((((......)))))........)))..))))))......(((..(.((((.....)))).)....))) ( -27.60, z-score = 0.31, R) >droEre2.scaffold_4784 8186773 74 - 25762168 GUGGCACUGCAGGAU---UCCCGCCUCAGCGUCCUGUUUCCAGGACCUG---CUCCAUAUGCCACCAUAUGCAUU-UUGGA------------------- ((((((..(((((..---...(((....)))(((((....)))))))))---)......))))))..........-.....------------------- ( -25.10, z-score = -2.11, R) >consensus GUGGCACUGCAGGAU___UCCCGCCUCAGCGUCCUGUUUCCAGGACCGG___CUCCAUCUGCCACCGCUCAUUCUGCUGCAUUCUGUGUGCUGACUUGCA (((((...((.((......)).))......((((((....))))))..............)))))................................... (-15.95 = -16.57 + 0.61)
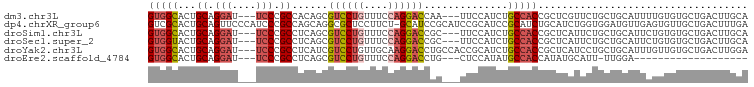
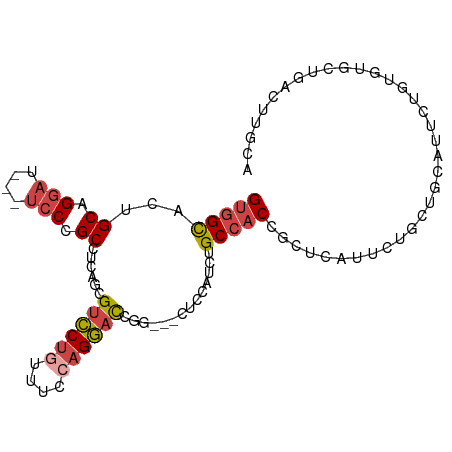
| Location | 5,501,984 – 5,502,090 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.16 |
| Shannon entropy | 0.60086 |
| G+C content | 0.52619 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -17.30 |
| Energy contribution | -19.10 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.882718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5501984 106 + 24543557 AACGAGCGGUGGCAGAUGGA---AUUGGUCCUGGAAACAGGACGCUGUGGCGGGAAUCCUGCAGUGCCACGCGUGCAUUGAACAUCGACAACAAAGCAAUUUCAGCUUA- ...((((.((((((......---....((((((....))))))......(((((...)))))..))))))...(((.(((...........))).)))......)))).- ( -37.50, z-score = -1.70, R) >droWil1.scaffold_180698 7311012 82 - 11422946 ------------------------AACAUGCGACUGACUACUCACUACACCAUGG---CCGCCG-ACAGAGCGUGCAUUGAACAUUGACAACAAAGCAAUUUUAGUUUUA ------------------------..((((....(((....)))......))))(---(((((.-...).))).))((((((.((((.(......))))))))))).... ( -10.50, z-score = 0.95, R) >dp4.chrXR_group6 5161454 100 - 13314419 ------CGGAUGCGGAUGCGGAUGCAGAAG--GAGCGCCUGCUGGCGGGAUGGGAA--CUGCAGUGCGACGCGUGCAUCGAACAUCGACAACAAAGCAAUUUUAGUUUUA ------.(.(((((..((((..(((((...--.....((((....)))).......--))))).)))).))))).).(((.....)))....(((((.......))))). ( -27.26, z-score = 0.79, R) >droSim1.chr3L 5017660 106 + 22553184 AAUGAGCGGUGGCAGAUGGA---AGCGGUCCUGGAAACAGGACGCUGAGGCGGGAAUCCUGCAGUGCCACGCGUGCAUUGAACAUCGACAACAAAGCAAUUUCAGCUUA- ((((.(((((((((......---((((.(((((....)))))))))...(((((...)))))..)))))).))).))))..............((((.......)))).- ( -41.80, z-score = -2.99, R) >droSec1.super_2 5438384 107 + 7591821 AAUGAGCGGUGGCAGAUGGA---AGCGGUCCUGGAAACAGGACGCUGAGGCGGGAAUCCUGCAGUACCACGCGUGCAUUGAACAUCGACAACAAAGCAAUUUCAGCUGAA .....((((((...((((.(---.(((((((((....))))))(.((..(((((...)))))..)).).))).).))))...))))).).....(((.......)))... ( -37.60, z-score = -2.10, R) >droYak2.chr3L 6064411 108 + 24197627 GAUGAGCGGUGGCAGAUGCGGUGGCAGGUCCUUGCAACAGGACGAUGAGGCGGGAAUCCUGCAGUGCCACGCGUGCAUUGAACAUCGACAACAAAGCAAUUUCAGCUA-- .....((((((...((((((((((((.(((((......)))))......(((((...)))))..))))))...))))))...))))).).....(((.......))).-- ( -37.20, z-score = -0.76, R) >droEre2.scaffold_4784 8186785 100 + 25762168 -----AUGGUGGCAUAUGGA---GCAGGUCCUGGAAACAGGACGCUGAGGCGGGAAUCCUGCAGUGCCACGCGUGCAUUGAACAUCGGCAACAAAGCAAUUUCAGCUA-- -----((((((((((.....---.(((((((((....)))))).)))..(((((...))))).))))))).)))((..((((.....((......))...))))))..-- ( -39.30, z-score = -2.61, R) >consensus _A_GAGCGGUGGCAGAUGGA___ACAGGUCCUGGAAACAGGACGCUGAGGCGGGAAUCCUGCAGUGCCACGCGUGCAUUGAACAUCGACAACAAAGCAAUUUCAGCUUA_ ........((((((.............((((((....))))))......((((.....))))..))))))((.(((.(((...........))).)))......)).... (-17.30 = -19.10 + 1.80)
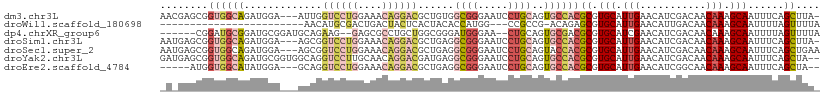
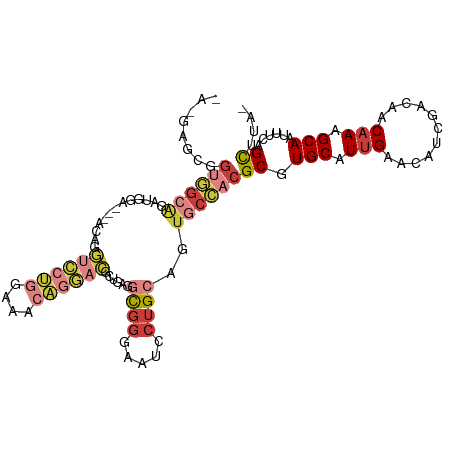
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:21 2011