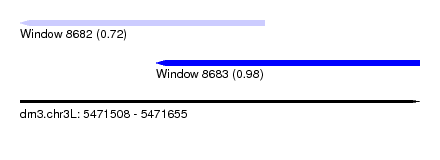
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,471,508 – 5,471,655 |
| Length | 147 |
| Max. P | 0.976997 |

| Location | 5,471,508 – 5,471,598 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 86.76 |
| Shannon entropy | 0.27599 |
| G+C content | 0.49696 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.04 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5471508 90 - 24543557 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCUGCAUGCGACGUGACCAGCAAC--UGACAUGUGCCACA .((((((((((((.(((((..(((((...)))))..)))........((((....))))))....))).)))))((--......)))))).. ( -28.30, z-score = -2.22, R) >droPer1.super_27 265126 87 - 1201981 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCAGCA---GACGUGACCAGCCCC--UGACAUGUGCCACA .((((.(((((((.(((((..(((((...)))))..)))..(((....(((....)))---....)))...))...--))))).)))))).. ( -27.20, z-score = -2.61, R) >dp4.chrXR_group6 4858613 87 + 13314419 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCAGCA---GACGUGACCAGCCCC--UGACAUGUGCCACA .((((.(((((((.(((((..(((((...)))))..)))..(((....(((....)))---....)))...))...--))))).)))))).. ( -27.20, z-score = -2.61, R) >droAna3.scaffold_13337 10212264 92 + 23293914 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGGCCCAGCACGAGACGUGACCAGCCCCCUUGGCAUGUGCCACA .((((.(((((((((.(((..(((((...)))))..)))..........(((....((((...))))....)))..))))))).)))))).. ( -31.50, z-score = -2.40, R) >droEre2.scaffold_4784 8156728 90 - 25762168 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCUGCAUCGGACGUGACCAGCAAC--UGUCAUGUGCCACA ..(((.(((....)))(((..(((((...)))))..)))..........)))........(((((((((.......--.))))))).))... ( -27.40, z-score = -1.59, R) >droYak2.chr3L 6032525 90 - 24197627 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCACAGCAUCCGACGUGACCAGCAAC--UGACAUGUGCCACA .((((((((((((..((((..(((((...)))))..)))........((((....))))..)...))).)))))((--......)))))).. ( -25.80, z-score = -1.41, R) >droSec1.super_2 5407871 90 - 7591821 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCUGCAUGCGACGUGACCAGCAAC--UGACAUGUGCCACA .((((((((((((.(((((..(((((...)))))..)))........((((....))))))....))).)))))((--......)))))).. ( -28.30, z-score = -2.22, R) >droSim1.chr3L 4985115 90 - 22553184 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCUGCAUGCGACGUGACCAGCAAC--UGACAUGUGCCACA .((((((((((((.(((((..(((((...)))))..)))........((((....))))))....))).)))))((--......)))))).. ( -28.30, z-score = -2.22, R) >droVir3.scaffold_13049 11398905 82 - 25233164 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUACGAGCUGGGC--AGACGUGACCGUC-----UAACAGCUGCC--- ..(((.((((((.((((((..(((((...)))))..)))..((......))))).)))--(((((....))))-----)...))).)))--- ( -28.70, z-score = -2.55, R) >droMoj3.scaffold_6680 15332672 82 + 24764193 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUACGGCCUGGGC--AGACGUGACCGUC-----UAACAGCUGCC--- ..(((.(((((.....(((..(((((...)))))..)))...........((....))--(((((....))))-----).))))).)))--- ( -27.90, z-score = -2.21, R) >droGri2.scaffold_15110 16615463 82 + 24565398 UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGGCCGGUGG--AGACGUGACUGUC-----UAACAGCUGCC--- ..(((.(((((.....(((..(((((...)))))..))).........(((.(((((.--...)...)))).)-----))))))).)))--- ( -26.00, z-score = -1.77, R) >consensus UUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACUAUCAAUUAUGCCCCAGCA__AGACGUGACCAGCAAC__UGACAUGUGCCACA ..(((.(((....)))(((..(((((...)))))..)))......(((((.............)))))..................)))... (-17.19 = -17.04 + -0.15)

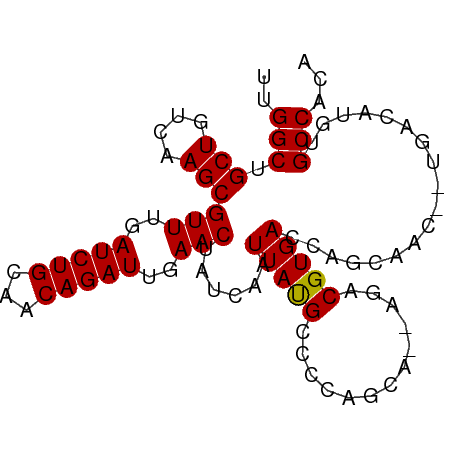
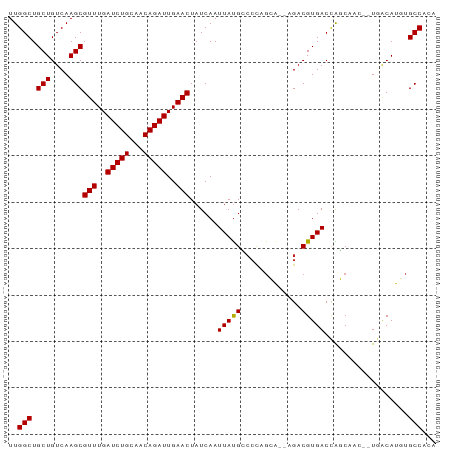
| Location | 5,471,558 – 5,471,655 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.13 |
| Shannon entropy | 0.57884 |
| G+C content | 0.44141 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5471558 97 - 24543557 ---AAAUGUUCCCACC-------UCUUUGGCCAAACUUGCUGCUGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ---.............-------.....((((((.....(.((((((......))))))..).....))))))(((....)))(((..(((((...)))))..))). ( -27.70, z-score = -2.04, R) >droPer1.super_27 265173 101 - 1201981 AUCAAAUUCCUCCAGA------GUCGUCAGAGCUCCUUUGGUGGGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ......((((.(((((------(..((....))..)))))).))))...........(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -31.80, z-score = -1.48, R) >dp4.chrXR_group6 4858660 101 + 13314419 AUCAAAUUCCUCCAGA------GUCGUCAGAGCUCCUUUGGUGGGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ......((((.(((((------(..((....))..)))))).))))...........(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -31.80, z-score = -1.48, R) >droAna3.scaffold_13337 10212316 107 + 23293914 AUCAAAUGUCUCCCCCUUUCCCUUUUUUUGUGAGUGCUGUUCUUGGACAAUUUUUCAGGUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ..............................(((..((.(((..(((((((((.....))))).))))..))).))..)))...(((..(((((...)))))..))). ( -20.10, z-score = 0.68, R) >droEre2.scaffold_4784 8156778 96 - 25762168 AUCAAAUGCUCCCACC-------UCUUUGGCUAAACUGG----UGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ...........(((((-------..(((....)))..))----)))...........(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -27.10, z-score = -1.91, R) >droYak2.chr3L 6032575 100 - 24197627 AUCACAUGCUUCCUCC-------UUUUUGGCCAAACUUGCUGCUGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ................-------.....((((((.....(.((((((......))))))..).....))))))(((....)))(((..(((((...)))))..))). ( -27.70, z-score = -1.78, R) >droSec1.super_2 5407921 97 - 7591821 ---AUAUGUUCCCAUC-------UCAUUGGCCAAGCUAGCUCCUGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ---.............-------...(((((..((((((((((((((......)))))...))...)))))))...)))))..(((..(((((...)))))..))). ( -24.20, z-score = -0.38, R) >droSim1.chr3L 4985165 100 - 22553184 AUCAUAUGUCCCCAUC-------UCAUUGGCCAAGUUUGCUGCUGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ......((((((((..-------....)))...(((.....))))))))........(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -28.20, z-score = -1.29, R) >droVir3.scaffold_13049 11398947 93 - 25233164 -----GUAUGAUGACC------AAU---CCAGAUAGCCCCAUGAAGGCAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU -----((.(((((.((------((.---.((((((((....((((((....)))))))))..)))))))))...).)))).))(((..(((((...)))))..))). ( -23.30, z-score = -0.59, R) >droMoj3.scaffold_6680 15332714 93 + 24764193 -----CUUAAAUGGCU------AAA---CCCAAUUGCCGCUGGAAGGCAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU -----......(((..------...---.)))((((((.......))))))......(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -27.60, z-score = -1.76, R) >droGri2.scaffold_15110 16615505 100 + 24565398 -CCCUGACUGACCACC------UAUGCCCAAAAUGGAGGCAUGAAGGCAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU -...(((..((...((------(((((((......).)))))..)))...))..)))(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -28.20, z-score = -1.24, R) >droWil1.scaffold_180698 7270539 83 + 11422946 ------------------------CUCAAAUGUGUCUUAGGGACAAACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU ------------------------........((((.....))))............(((((((..((.....)).)))))))(((..(((((...)))))..))). ( -22.70, z-score = -1.48, R) >consensus _UCAAAUGUCCCCACC_______UCUUUGGCGAAACUUGCUGGUGGACAAUUUUUCAGCUUGAUCUGUUGGCUGCUGUCAAGCGUUUGAUCUGCAACAGAUUGAACU .........................................................(((((((..((.....)).)))))))(((..(((((...)))))..))). (-18.76 = -18.84 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:18 2011