| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,445,766 – 5,445,856 |
| Length | 90 |
| Max. P | 0.749644 |

| Location | 5,445,766 – 5,445,856 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 71.02 |
| Shannon entropy | 0.55191 |
| G+C content | 0.37288 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -9.13 |
| Energy contribution | -8.61 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5445766 90 + 24543557 UUCGGAUUUUUUUUUCGUUGAGUU--UUCCACUUCGUAUAAGAGUUUUCCU-GCGCGUGCGUGUG-AGUUAAUAUUUUGUGGAGAGUUCAACUC---- ................(((((..(--((((((.((......))(((..(..-((((....)))).-.)..))).....)))))))..)))))..---- ( -22.30, z-score = -2.14, R) >droAna3.scaffold_13337 10195103 89 - 23293914 -UUCGGUAACCAUUUCGUUAAGUU--UU-CAAUCCGUAUAAGAGUUUUUUAAGCGCAUGCGUGAA-AAUUAAUAAUUUUUGGAGAAUUCCGCGC---- -..(((...(((....((((((((--((-((...(((((....(((.....)))..)))))))))-))))..))))...)))......)))...---- ( -12.50, z-score = 0.89, R) >droSim1.chr3L 17619557 89 - 22553184 UUCGGAUUUUUGUU-CGUUGAGUU--UUCCACUUCGUAUAAGAGUUUUCCU-GCGCGUGCGUGUU-AGUUAAUAUUUUGUGGAGAGUUCAACUC---- ...((((....)))-)(((((..(--((((((.((......))(((..(..-((((....)))).-.)..))).....)))))))..)))))..---- ( -23.00, z-score = -2.62, R) >droSec1.super_2 5390884 89 + 7591821 UUCGGAUUUUUGUU-CGUUGAGUU--UUCCACUUCGUAUAAGAGUUUUCCU-GCGCGUGCGUGUG-AGUUUAUAUUUUGUGGAGAGUUCAACUC---- ...((((....)))-)(((((..(--((((((...(((((((....(((.(-(((....)))).)-)))))))))...)))))))..)))))..---- ( -25.80, z-score = -3.35, R) >droYak2.chr3L 6015012 89 + 24197627 UUCGGAUUUUUGUU-CGUUGAGUU--UUCCACUUCGUAUAAGAGUUUUUCU-GCGCGUGCGUUUG-AGUUAAUAUUUUGUGGAGAGUUCAACUC---- ...((((....)))-)(((((..(--(((((((((......)))...(((.-(((....)))..)-))..........)))))))..)))))..---- ( -22.10, z-score = -2.04, R) >droEre2.scaffold_4784 8140020 89 + 25762168 UUCCGAUUUCUGUU-CGUUGAGUU--UUUCACAUUGUAUAAGAGUUUUCCU-GCGCGUGCGUGUG-AGUUAAUAUUUUGUGGAGAGUUCAGCUC---- ..............-.(((((..(--(((((((..((((.......(((.(-(((....)))).)-))...))))..))))))))..)))))..---- ( -21.40, z-score = -1.80, R) >anoGam1.chr3L 38199616 97 - 41284009 -UAAAAAAUUAAUAUCAAAGGAUUAAUAUCGAACUUUCCAAAGUUUUUCAUGUUUUGGUUGUAAACAAACGAUCUCUAUUUCAGAUACCACUGCAGUC -..................((.....((((((((((....))))))..........((((((......)))))).........))))))......... ( -8.90, z-score = 1.11, R) >consensus UUCGGAUUUUUGUU_CGUUGAGUU__UUCCACUUCGUAUAAGAGUUUUCCU_GCGCGUGCGUGUG_AGUUAAUAUUUUGUGGAGAGUUCAACUC____ ................((((((....(((((((((......)))........((((....))))..............))))))..))))))...... ( -9.13 = -8.61 + -0.52)

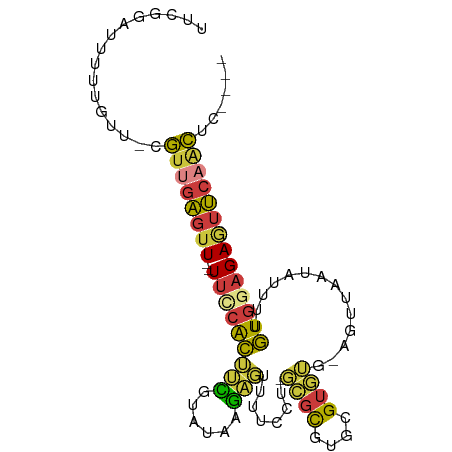
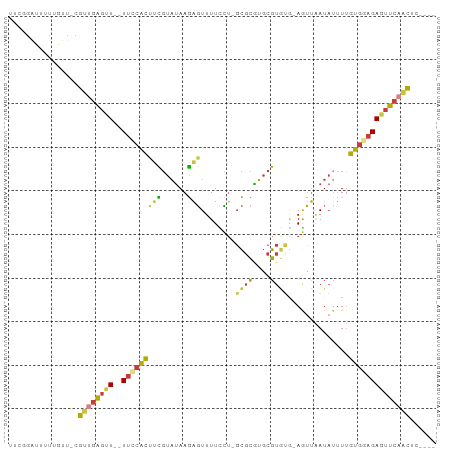
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:14 2011