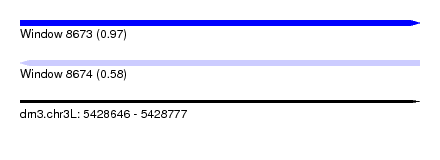
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,428,646 – 5,428,777 |
| Length | 131 |
| Max. P | 0.974364 |

| Location | 5,428,646 – 5,428,777 |
|---|---|
| Length | 131 |
| Sequences | 11 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 75.01 |
| Shannon entropy | 0.52722 |
| G+C content | 0.52131 |
| Mean single sequence MFE | -35.91 |
| Consensus MFE | -27.36 |
| Energy contribution | -27.35 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
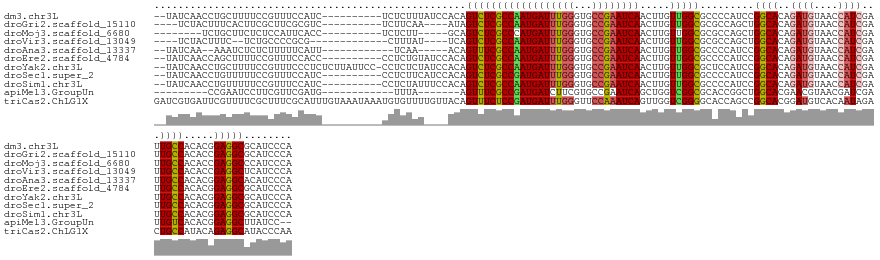
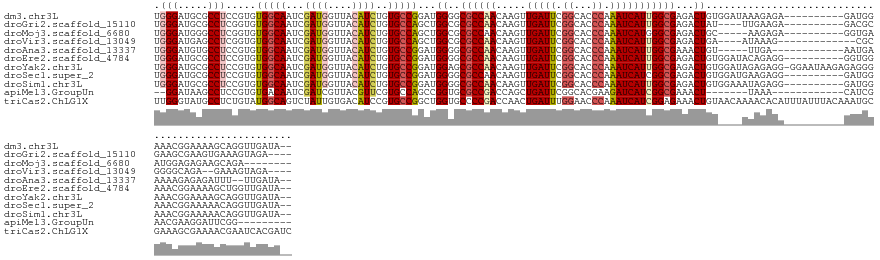
>dm3.chr3L 5428646 131 + 24543557 --UAUCAACCUGCUUUUCCGUUUCCAUC----------UCUCUUUAUCCACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCA --........(((.(((((((.......----------.........................(((..(((((((((.........))))))))).))).((((..((((....))))...))))..))))))).)))..... ( -36.30, z-score = -1.88, R) >droGri2.scaffold_15110 7979056 125 + 24565398 ----UCUACUUUCACUUCGCUUCGCGUC----------UCUUCAA----AUAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCGCAUCCCA ----...................(((((----------((.....----...(.(.((((((((((((((...)))))))).....)))))).).).(.(((((..((((....))))...))))))...)))))))...... ( -38.90, z-score = -1.98, R) >droMoj3.scaffold_6680 9464392 120 + 24764193 --------UCUGCUUCUCUCCAUUCACC----------UCUCUU-----GCAGUCUCGCCCAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCCCAUCCCA --------..................((----------((....-----((((((..(((..((((((((...))))))))....(((((....))))).)))...((((....))))))))))......))))......... ( -33.70, z-score = -0.78, R) >droVir3.scaffold_13049 11361757 120 + 25233164 ----UCUACUUUC--UCUGCCCCGCG-------------CUUUAU----UCAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCGCCAGCUGGCACAGAUGUAACCAUCGAUUGCCACACCGAGGCUCAUCCCA ----.........--...(((.((.(-------------......----.(((((..((((.((((((((...))))))))....(((((....)))))))))...((((....))))))))).....))).)))........ ( -32.60, z-score = -0.36, R) >droAna3.scaffold_13337 10178457 122 - 23293914 --UAUCAA--AAAUCUCUCUUUUUCAUU------------UCAA-----ACAGUUUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCACAUCCCA --......--..................------------....-----........(((........(((((((((.........)))))))))..(((((((..((((....))))...))))....))))))........ ( -31.20, z-score = -1.51, R) >droEre2.scaffold_4784 8122021 131 + 25762168 --UAUCAACCAGCUUUUCCGUUUCCACC----------CCUCUGUAUCCACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCA --.........((.(((((((.......----------...((((....))))..........(((..(((((((((.........))))))))).))).((((..((((....))))...))))..))))))).))...... ( -37.30, z-score = -1.73, R) >droYak2.chr3L 5997706 140 + 24197627 --UAUCAACCUGCUUUUCCGUUUCCCUCUCUUAUUCC-CCUCUCUAUCCACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCUCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCA --........(((.(((((((................-..................((((((((((((((...)))))))).....))))))........((((..((((....))))...))))..))))))).)))..... ( -35.60, z-score = -1.76, R) >droSec1.super_2 5373795 131 + 7591821 --UAUCAACCUGUUUUUCCGUUUCCAUC----------CCUCUUCAUCCACAGUCUCGCCGAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCA --...((((..(((.(((.((......(----------((...(((((..(......)..)))))...))).)).)))..)))..))))(((((((....((((..((((....))))...))))....)).)))))...... ( -38.10, z-score = -1.89, R) >droSim1.chr3L 4943248 131 + 22553184 --UAUCAACCUGUUUUUCCGUUUCCAUC----------CCUCUAUUUCCACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCA --................(((((((...----------.........................(((..(((((((((.........))))))))).))).((((..((((....))))...))))....)))))))....... ( -34.80, z-score = -1.40, R) >apiMel3.GroupUn 202380634 113 - 399230636 ---------CCGAAUCCUUCGUUCGAUG------------UUUA-------AGUUUCGCCGAUGAUCUUCGUGCCGAAUCAGCUGGUCGGCGCACCGGCUGGCACGAACGUAACGAUCGAUUGUCACACGGAGGCUUAUCC-- ---------......((((((((((...------------....-------.(.((((((((......))).).)))).).((..(((((....)))))..)).)))))((.((((....)))).))..))))).......-- ( -36.00, z-score = -0.31, R) >triCas2.ChLG1X 7529755 143 - 8109244 GAUCGUGAUUCGUUUUCGCUUUCGCAUUUGUAAAUAAAUGUGUUUUGUUACAGUUUCUCCGAUGAUUUGGGUUCCAAAUCAGUUGGUCGGGGCACCAGCCGGCACGGAUGUCACAAUAGACUGCCAUACAGAGGCAUACCCAA ((..((((.......))))..))((.((((((.......((.(.((((.(((.....((((.((((((((...))))))))((((((.((....)).)))))).))))))).)))).).)).....)))))).))........ ( -40.50, z-score = -1.02, R) >consensus __UAUCAACCUGCUUUUCCGUUUCCAUC__________CCUCUAU____ACAGUCUCGCCAAUGAUUUGGGUGCCGAAUCAACUUGUUGGCGCCCCAUCCGGCACAGAUGUAACCAUCGAUUGCCACACGGAGGCGCAUCCCA ....................................................((((((((((((((((((...)))))))).....))))).........((((..((((....))))...)))).....)))))........ (-27.36 = -27.35 + -0.01)
| Location | 5,428,646 – 5,428,777 |
|---|---|
| Length | 131 |
| Sequences | 11 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 75.01 |
| Shannon entropy | 0.52722 |
| G+C content | 0.52131 |
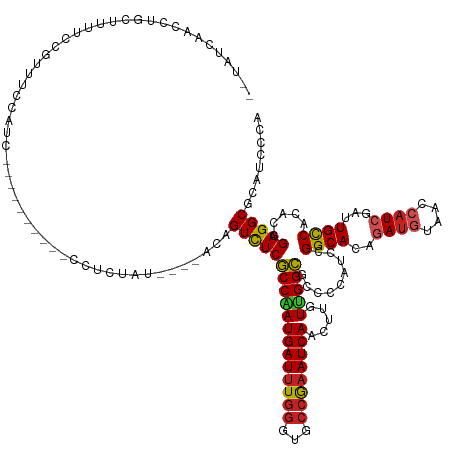
| Mean single sequence MFE | -42.61 |
| Consensus MFE | -20.41 |
| Energy contribution | -20.71 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
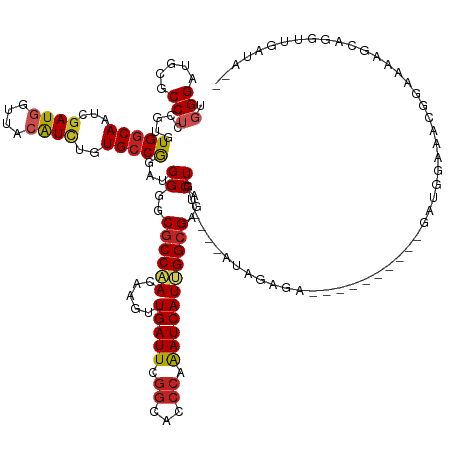
>dm3.chr3L 5428646 131 - 24543557 UGGGAUGCGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGGGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUGUGGAUAAAGAGA----------GAUGGAAACGGAAAAGCAGGUUGAUA-- ......(((((.((((.(((((...((((....))))..))))).)))))))))((((..(((..((((....(((..(((((.(((....))).))).....))----------..)))...))))..)))..))))...-- ( -44.70, z-score = -1.62, R) >droGri2.scaffold_15110 7979056 125 - 24565398 UGGGAUGCGCCUCGGUGUGGCAAUCGAUGGUUACAUCUGUGCCAGCUGGCGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUAU----UUGAAGA----------GACGCGAAGCGAAGUGAAAGUAGA---- ......(((((..(((.(((((...((((....))))..)))))))))))))(((((.....(((((.((...)).))))))))))....((((----((.....----------..(((...)))......)))))).---- ( -38.52, z-score = 0.02, R) >droMoj3.scaffold_6680 9464392 120 - 24764193 UGGGAUGGGCCUCGGUGUGGCAAUCGAUGGUUACAUCUGUGCCAGCUGGCGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUGGGCGAGACUGC-----AAGAGA----------GGUGAAUGGAGAGAAGCAGA-------- ........(((((..((((((....((((....)))).(((((....))))))))....((((...(((...((((.....)))))))))))))-----)...))----------))).................-------- ( -35.60, z-score = 1.01, R) >droVir3.scaffold_13049 11361757 120 - 25233164 UGGGAUGAGCCUCGGUGUGGCAAUCGAUGGUUACAUCUGUGCCAGCUGGCGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUGA----AUAAAG-------------CGCGGGGCAGA--GAAAGUAGA---- ........((((((((.((((....((((....)))).(((((....)))))))))))..(((.(((((((.(((((....)))).).).))))----))..))-------------).))))))...--.........---- ( -40.50, z-score = -0.69, R) >droAna3.scaffold_13337 10178457 122 + 23293914 UGGGAUGUGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGGGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAAACUGU-----UUGA------------AAUGAAAAAGAGAGAUUU--UUGAUA-- ((((..(((((.((((.(((((...((((....))))..))))).)))))))))......((((...)))).))))..(((((..((((....)-----))).------------))))).............--......-- ( -38.90, z-score = -1.51, R) >droEre2.scaffold_4784 8122021 131 - 25762168 UGGGAUGCGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGGGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUGUGGAUACAGAGG----------GGUGGAAACGGAAAAGCUGGUUGAUA-- ......(((((.((((.(((((...((((....))))..))))).)))))))))((((.((((..((((.(((((...((......))..((.((....)).)))----------))))....))))..)))).))))...-- ( -52.30, z-score = -2.80, R) >droYak2.chr3L 5997706 140 - 24197627 UGGGAUGCGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGAGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUGUGGAUAGAGAGG-GGAAUAAGAGAGGGAAACGGAAAAGCAGGUUGAUA-- ......((((.(((((.(((((...((((....))))..))))).)))))))))((((..(((..(((....(((...(((((.(((....))).))).))...)-))..........(....))))..)))..))))...-- ( -47.30, z-score = -2.11, R) >droSec1.super_2 5373795 131 - 7591821 UGGGAUGCGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGGGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUCGGCGAGACUGUGGAUGAAGAGG----------GAUGGAAACGGAAAAACAGGUUGAUA-- ......(((((.((((.(((((...((((....))))..))))).)))))))))((((..(((..((((.(((((...(((((.(((....))).)))))...))----------).))....))))..)))..))))...-- ( -52.90, z-score = -3.39, R) >droSim1.chr3L 4943248 131 - 22553184 UGGGAUGCGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGGGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUGUGGAAAUAGAGG----------GAUGGAAACGGAAAAACAGGUUGAUA-- ......(((((.((((.(((((...((((....))))..))))).)))))))))((((..(((..((((.(((((...((......))..((((....)))).))----------).))....))))..)))..))))...-- ( -47.60, z-score = -2.36, R) >apiMel3.GroupUn 202380634 113 + 399230636 --GGAUAAGCCUCCGUGUGACAAUCGAUCGUUACGUUCGUGCCAGCCGGUGCGCCGACCAGCUGAUUCGGCACGAAGAUCAUCGGCGAAACU-------UAAA------------CAUCGAACGAAGGAUUCGG--------- --......(((.....(((((........))))).(((((((((((.(((......))).))).....)))))))).......)))......-------....------------...(((((....).)))).--------- ( -32.40, z-score = 0.56, R) >triCas2.ChLG1X 7529755 143 + 8109244 UUGGGUAUGCCUCUGUAUGGCAGUCUAUUGUGACAUCCGUGCCGGCUGGUGCCCCGACCAACUGAUUUGGAACCCAAAUCAUCGGAGAAACUGUAACAAAACACAUUUAUUUACAAAUGCGAAAGCGAAAACGAAUCACGAUC ..(((((((((...((((((..(((......)))..)))))).)))..))))))((......((((((((...))))))))(((.......(((((..............)))))..(((....)))....)))....))... ( -37.94, z-score = -1.38, R) >consensus UGGGAUGCGCCUCCGUGUGGCAAUCGAUGGUUACAUCUGUGCCGGAUGGGGCGCCAACAAGUUGAUUCGGCACCCAAAUCAUUGGCGAGACUGU____AUAGAGA__________GAUGGAAACGGAAAAGCAGGUUGAUA__ .(((.....))).....(((((...((((....))))..)))))...((..((((((.....(((((.((...)).)))))))))))...))................................................... (-20.41 = -20.71 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:11 2011