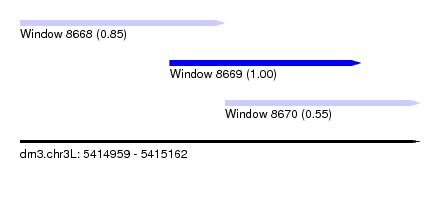
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,414,959 – 5,415,162 |
| Length | 203 |
| Max. P | 0.998945 |

| Location | 5,414,959 – 5,415,063 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.66851 |
| G+C content | 0.44002 |
| Mean single sequence MFE | -19.39 |
| Consensus MFE | -7.30 |
| Energy contribution | -7.45 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.851423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5414959 104 + 24543557 -AGAUGCGGUCUCAACUUGUUU--CCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGGCACUU-CAACUUC-------------UCCCCCUUUUUCGCAUCUUACCUUU -((((((((.............--...((.(((.((((((.....)))))).))).))..........((.....-.......-------------....))....))))))))....... ( -17.89, z-score = -1.59, R) >droSim1.chr3L 4932400 110 + 22553184 -AGAUGCGGUCUCAACUUGUUU--CCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUU-CAACUUCGUCUUC-------UCCCCCUUUUUCGCAUCUUACCUUU -((((((((.............--...((.(((.((((((.....)))))).))).))..........(((....-.......)))...-------..........))))))))....... ( -20.40, z-score = -2.80, R) >droSec1.super_2 5360293 110 + 7591821 -AGAUGCGGUCUCAACUUGUUU--CCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUU-CAACUUCGUAUUC-------UCCUCCUUUUUCGCAUCUUACCUUU -((((((((.............--...((.(((.((((((.....)))))).))).)).......(((((.....-.....)))))...-------..........))))))))....... ( -21.60, z-score = -3.12, R) >droYak2.chr3L 5983048 117 + 24197627 -AGAUGCGGUCUCAACUUGUUU--CCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUU-CCACUUCGUCCUCGCUCUCUUCGUCCUUGUUCACAUCUUACCUUU -(((((.((...(((.......--...((.(((.((((((.....)))))).))).))........((((.((..-.......))..))))...........))).)).)))))....... ( -17.40, z-score = -0.74, R) >droEre2.scaffold_4784 8108211 103 + 25762168 -AGACGCGGUCUCAACUUGUUU--CCCAAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUU-CGUCUUCUCCC--------------CUUUUUCACAUCUUACCUUU -(((((.(((.((.....((..--......(((.((((((.....)))))).)))........))...)).))).-)))))......--------------.................... ( -15.39, z-score = -1.80, R) >droAna3.scaffold_13337 10165410 106 - 23293914 ----AACUGCCUCGACUUGGUU--CCAGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGCCACUC-CAGCUCCGAUUCG--------CAGCCCUUCCCCCAGAUGUCCUGG ----....((..(((.((((..--...((.(((.((((((.....)))))).))).))........((.......-..)).)))).)))--------..)).......((((.....)))) ( -21.40, z-score = -0.68, R) >droWil1.scaffold_180698 7202171 85 - 11422946 --GGUUAAGUUU--ACUUGGUU----CGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUUUCCACUACCUUU-------------CCUUUU--------------- --((..(((((.--.(.(((((----.((.(((.((((((.....)))))).))).)).)))))..)..).)))).)).........-------------......--------------- ( -13.90, z-score = -1.12, R) >droPer1.super_27 201722 111 + 1201981 AGCUGUGUGUCUCAGCGCCUUU-CCCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUC-CGGCACUGUCGCGCA------CCUUCCCUCUUGUGUAUCUGCU-- ....((((((..(((.(((...-....((.(((.((((((.....)))))).))).))..........(......-)))).)))..)))))------).....................-- ( -25.80, z-score = -1.15, R) >dp4.chrXR_group6 8478325 112 + 13314419 AGCUGUGUGUCUCAGCGUCUUUUCCCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUC-CGGCACUGUCGCGCA------CCUUCCCUCUUGUGUAUCUGCU-- .((((.(((((.(((.(((........)))....((((((.....)))))).............))).)))))..-))))......(((((------..........))))).......-- ( -25.80, z-score = -1.28, R) >droVir3.scaffold_13049 11348387 83 + 25233164 ----AUCAGUUUUAGCUUCUUU--UCCGAUAUUUGCAUGUCAAACGCGUGGUAAUUUUAUAUUACUGCGACACUC--GAUACCGUCUCUGA------------------------------ ----.((((........((...--...(((((....)))))....(((..(((((.....)))))..)).)....--))........))))------------------------------ ( -14.89, z-score = -0.77, R) >droGri2.scaffold_15110 7965948 94 + 24565398 ----AUCAGUCGCAACUUAUUU--UUCGAUAUUUGCAUGUCAAACGCGUGGUAAUUUUAUAUUACUGCGACACUC--GACUUGGCUU-------------CCCUUCCCUGACUCU------ ----...((((((((..((((.--...)))).))))..(((.....((..(((((.....)))))..))......--))).......-------------.........))))..------ ( -18.80, z-score = -1.96, R) >consensus _AGAUGCGGUCUCAACUUGUUU__CCCGAUAUUUGCAUGUCAAACGCGUGCUAAUUUCAAAUUACUGCGACACUU_CGACUUCGUCU__________CC_CCUUUUUCGCAUCUUACCU__ ...........................((.(((.((((((.....)))))).))).))............................................................... ( -7.30 = -7.45 + 0.14)
| Location | 5,415,035 – 5,415,132 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 62.34 |
| Shannon entropy | 0.79838 |
| G+C content | 0.44013 |
| Mean single sequence MFE | -12.25 |
| Consensus MFE | -7.07 |
| Energy contribution | -6.76 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.998945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
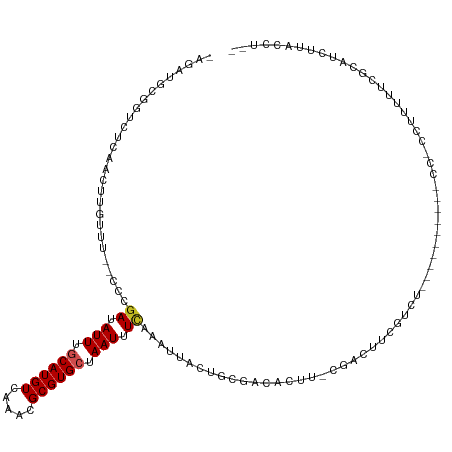
>dm3.chr3L 5415035 97 + 24543557 ------------UUCUCCCCCUUUUUCGC-AUCUUACCUUUUUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGUGUCAUUUUAUAAA ------------...............((-((...........)))).............((......))....(((((((..((.((((.--....))))))..))))))) ( -10.50, z-score = -1.84, R) >droSim1.chr3L 4932477 102 + 22553184 -------UCGUCUUCUCCCCCUUUUUCGC-AUCUUACCUUUUUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAA -------....................((-((...........)))).............((......))....(((((((..((.((((.--....))))))..))))))) ( -12.40, z-score = -2.14, R) >droSec1.super_2 5360370 102 + 7591821 -------UCGUAUUCUCCUCCUUUUUCGC-AUCUUACCUUUUUAUGCCCCGUCUCCACCAGCUAAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUCUAUAAA -------(((((...............((-((...........)))).............((......))......)))))..((.((((.--....))))))......... ( -10.00, z-score = -1.42, R) >droYak2.chr3L 5983125 109 + 24197627 UCGUCCUCGCUCUCUUCGUCCUUGUUCAC-AUCUUACCUUUUUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU--UGCUGCGUCAUUUUAUAAA .............................-.............((((....((..(....)..))...))))..(((((((..((.((((.--....))))))..))))))) ( -12.10, z-score = -0.33, R) >droEre2.scaffold_4784 8108288 95 + 25762168 --------------UCUCCCCUUUUUCAC-AUCUUACCUUUUUAUGCCCCGUCUCCACCAGCCGAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAA --------------...............-.............((((..((.((.....)).))....))))..(((((((..((.((((.--....))))))..))))))) ( -12.00, z-score = -2.86, R) >droAna3.scaffold_13337 10165484 100 - 23293914 --CCGAUUCGCAGCCCUUCCCCCAGAUGU-CCUGGUUCCCCUCCGGCCCACCCAC-------CCGAACGCAUUAUUUAUGACUCGUACGCU--UUCUGCGUCAUUUUAUAAA --.......((..........((((....-.))))........(((.........-------)))...))....(((((((...(.((((.--....)))))...))))))) ( -15.10, z-score = -0.42, R) >droWil1.scaffold_180698 7202238 81 - 11422946 ------------------------UCCACUACCUUUCCUUUUU----UCCGCC--CACAAGUCGAAG-GCAUUAUUUAUGACUUGUACGCUGCUUCUGCGUCAUUUUAUGAA ------------------------...................----...(((--..(.....)..)-))....(((((((..((.((((.......))))))..))))))) ( -13.10, z-score = -1.44, R) >droPer1.super_27 201801 104 + 1201981 --CUGUCGCGCACCUUCCCUCUUGUGUAU-CUGCUCCCUGCUCCACCUCCCCCUC---CUCUCUGAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAA --.....((((((..........))))..-..((.....))..............---..........))....(((((((..((.((((.--....))))))..))))))) ( -15.40, z-score = -3.43, R) >dp4.chrXR_group6 8478405 107 + 13314419 --CUGUCGCGCACCUUCCCUCUUGUGUAU-CUGCUCCCUGCUCCACCUCCCCCCCUCCCUCCCUGAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAA --.....((((((..........))))..-..((.....))...........................))....(((((((..((.((((.--....))))))..))))))) ( -15.40, z-score = -3.74, R) >droVir3.scaffold_13049 11348458 74 + 25233164 ------------------------------------UACCGUCUCUGAGCCCUUCGCGUUGCUCGAACACAUUAUUUAUGACUUGUACGUU--UUCCGUGCUAUUUUAUAAA ------------------------------------....((.((.((((..........))))))))......(((((((...(((((..--...)))))....))))))) ( -11.60, z-score = -1.02, R) >droGri2.scaffold_15110 7966020 86 + 24565398 ------------------------UUGGCUUCCCUUCCCUGACUCUUUGCAUCUCUCAGUGCUCGAACGCAUUAUUUAUGACUUGUACGUU--UUCCGUGUUAUUUUAUAAA ------------------------...(((((.(......).......((((......))))..))).)).((((..(((((....(((..--...))))))))...)))). ( -7.20, z-score = 1.04, R) >consensus ______________CUCCCCCUUUUUCAC_AUCUUACCUUUUUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU__UUCUGCGUCAUUUUAUAAA ..........................................................................(((((((..((.((((.......))))))..))))))) ( -7.07 = -6.76 + -0.30)
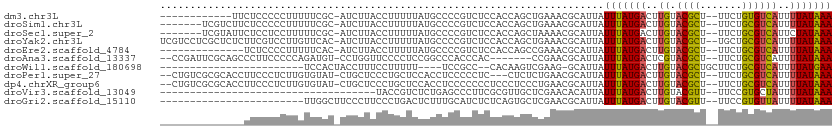
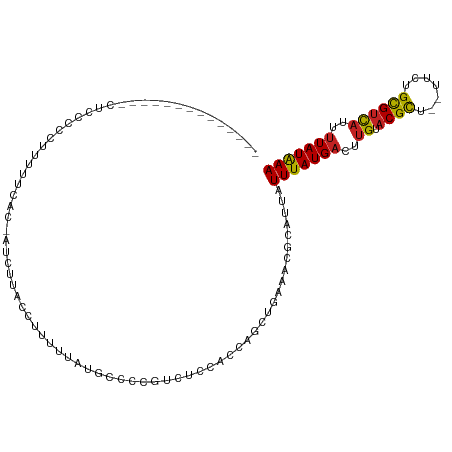
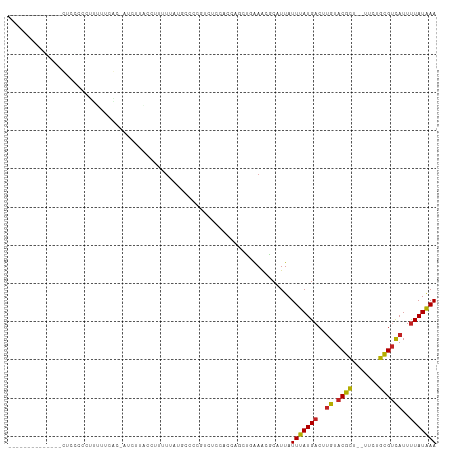
| Location | 5,415,063 – 5,415,162 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.08 |
| Shannon entropy | 0.63151 |
| G+C content | 0.43263 |
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -7.11 |
| Energy contribution | -6.69 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5415063 99 + 24543557 ------UUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGUGUCAUUUUAUAAACAUUUUCCAAA-CAU-------GUUCGGCGACCUCGGA ------.....((..(((.((....((......))....(((((((..((.((((.--....))))))..)))))))...........-...-------....)).)))...)). ( -17.50, z-score = -0.75, R) >droSim1.chr3L 4932510 99 + 22553184 ------UUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAACAUUUUCCAAA-CAU-------GUUCGGCGACCUCGGA ------.....((..(((.((....((......))....(((((((..((.((((.--....))))))..)))))))...........-...-------....)).)))...)). ( -19.40, z-score = -1.41, R) >droSec1.super_2 5360403 99 + 7591821 ------UUAUGCCCCGUCUCCACCAGCUAAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUCUAUAAACAUUUUCCAAA-CAU-------GUUCGGCGACCUCGGA ------.....((..(((.((....((......))....((((((...((.((((.--....))))))...))))))...........-...-------....)).)))...)). ( -16.80, z-score = -0.91, R) >droYak2.chr3L 5983165 106 + 24197627 ------UUAUGCCCCGUCUCCACCAGCUGAAACGCAUUAUUUAUGACUUGUACGCU--UGCUGCGUCAUUUUAUAAACAUUUUCCAAA-CAUUCUCCAUGUUCUGCGACCUCGGG ------......((((((..(....)..))..((((...(((((((..((.((((.--....))))))..))))))).........((-(((.....))))).))))....)))) ( -20.20, z-score = -1.65, R) >droEre2.scaffold_4784 8108314 99 + 25762168 ------UUAUGCCCCGUCUCCACCAGCCGAAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAACAUUUUCCAAA-CAU-------GUUCGGCGACCUCGGA ------.......(((.........((((((........(((((((..((.((((.--....))))))..)))))))(((........-.))-------))))))).....))). ( -20.84, z-score = -1.91, R) >droAna3.scaffold_13337 10165516 89 - 23293914 ------UUCCCCUCCGGCCCAC-CCACCCGAACGCAUUAUUUAUGACUCGUACGCU--UUCUGCGUCAUUUUAUAAACAUUUUCCAAA-CAU-------GUU--CCGA------- ------........(((..((.-.....(((...(((.....)))..))).((((.--....))))......................-..)-------)..--))).------- ( -11.30, z-score = -1.38, R) >droWil1.scaffold_180698 7202256 93 - 11422946 -----------UUCCGCCCA---CAAGUCGAAGGCAUUAUUUAUGACUUGUACGCUGCUUCUGCGUCAUUUUAUGAACAUUUUUCAAAAUAU-----GUUUUCAGCAUCAUA--- -----------.........---...((.((((((((..(((((((..((.((((.......))))))..))))))).((((....))))))-----)))))).))......--- ( -17.00, z-score = -1.16, R) >droPer1.super_27 201833 99 + 1201981 CCCUGCUCCACCUCCCCCUC---CUCUCUGAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAACAUUUUCGAAA-CAU-------GUUGGCCAGAAGG--- ...................(---((.((((..(((((..(((((((..((.((((.--....))))))..)))))))(......)...-.))-------).))..)))))))--- ( -16.80, z-score = -1.69, R) >dp4.chrXR_group6 8478437 102 + 13314419 CCCUGCUCCACCUCCCCCCCUCCCUCCCUGAACGCAUUAUUUAUGACUUGUACGCU--UUCUGCGUCAUUUUAUAAACAUUUUCGAAA-CAU-------GUUGGCCAGAAGG--- ......................(((..(((..(((((..(((((((..((.((((.--....))))))..)))))))(......)...-.))-------).))..))).)))--- ( -15.00, z-score = -1.12, R) >droVir3.scaffold_13049 11348470 93 + 25233164 ----------GCCCUUCGCGUUG---CUCGAACACAUUAUUUAUGACUUGUACGUU--UUCCGUGCUAUUUUAUAAACAUUUUUCAAAAUAUGCUGCCCGCCCGCCAA------- ----------.......(((..(---(............(((((((...(((((..--...)))))....)))))))(((..........)))......)).)))...------- ( -11.00, z-score = 0.32, R) >droMoj3.scaffold_6680 15277810 99 - 24764193 ----------GCCCCUCCCUUUCACCCUCGAACGCAUUAUUUAUGACUUGUACGUU--UUCCGUGUUAUUUUAUAAACAUUUUUCAAAAUAUGCUGCCCGCGCGAGUCUGC---- ----------..................................((((((..((..--(..((((((....................))))))..)..))..))))))...---- ( -12.65, z-score = -0.58, R) >droGri2.scaffold_15110 7966042 96 + 24565398 --------UUGCAUCUCUCAGUG---CUCGAACGCAUUAUUUAUGACUUGUACGUU--UUCCGUGUUAUUUUAUAAACAUUUUUCAAAAUAUGCUGCCCGCCCGUCCGA------ --------..((((.....((((---(......))))).(((((((....((((..--...)))).....))))))).............))))...............------ ( -10.50, z-score = 1.01, R) >consensus ______UUAUGCCCCGCCCCCACCAGCUCGAACGCAUUAUUUAUGACUUGUACGCU__UUCUGCGUCAUUUUAUAAACAUUUUCCAAA_CAU_______GUUCGGCGACCUC___ .......................................(((((((..((.((((.......))))))..)))))))...................................... ( -7.11 = -6.69 + -0.42)
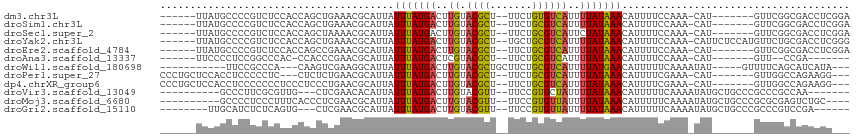

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:07 2011