
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,412,921 – 5,413,014 |
| Length | 93 |
| Max. P | 0.879930 |

| Location | 5,412,921 – 5,413,014 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.39129 |
| G+C content | 0.47150 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.15 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
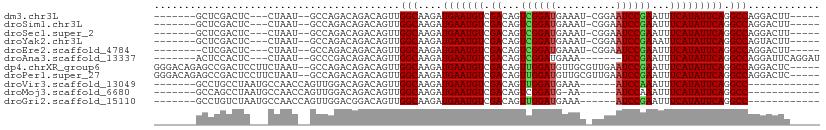
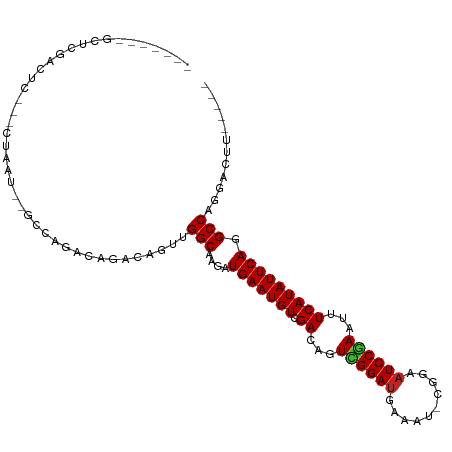
>dm3.chr3L 5412921 93 - 24543557 -------GCUCGACUC---CUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAAU-CGGAAUCCGAAUUUCAUAUUCAGGCCAGGACUU----- -------.......((---((...--(((.....(((.(((((((.......))))))).)))..(((((((-(((...))).))))))).....))).))))...----- ( -27.00, z-score = -1.56, R) >droSim1.chr3L 4930338 93 - 22553184 -------GCUCGACUC---CUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAAU-CGGAAUCCGAAUUUCAUAUUCAGGCCAGGACUU----- -------.......((---((...--(((.....(((.(((((((.......))))))).)))..(((((((-(((...))).))))))).....))).))))...----- ( -27.00, z-score = -1.56, R) >droSec1.super_2 5358400 93 - 7591821 -------GCUCGACUC---CUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAAU-CGGAAUCCGAAUUUCAUAUUCAGGCCAGGACUU----- -------.......((---((...--(((.....(((.(((((((.......))))))).)))..(((((((-(((...))).))))))).....))).))))...----- ( -27.00, z-score = -1.56, R) >droYak2.chr3L 5981064 93 - 24197627 -------GCUCGACUC---CUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAAU-CGGAAUCCGAAAUUCAUAUUCAGGCCAGUACUU----- -------.....((((---((..(--(((((........))))))....((((((.((...((((((.....-....))))))...)))))))))))..)))....----- ( -23.20, z-score = -0.80, R) >droEre2.scaffold_4784 8106210 92 - 25762168 --------CUCGACUC---CUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAAU-CGGAAUCCGAAUUUCAUAUUCAGGCCAGGACUU----- --------......((---((...--(((.....(((.(((((((.......))))))).)))..(((((((-(((...))).))))))).....))).))))...----- ( -27.00, z-score = -1.74, R) >droAna3.scaffold_13337 10163308 92 + 23293914 -------ACUCCACUC---CUAAU--GCCCGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAA-------UCCGAAUUUCAUAUUCAGGCCAGGAUUCAGGAU -------..(((..((---((..(--(((.(((.....)))))))...(((((((.((...((((((...)-------)))))...)))))))))....))))....))). ( -25.60, z-score = -1.36, R) >dp4.chrXR_group6 8475770 104 - 13314419 GGGACAGAGCCGACUCCUUCUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUUGGAUGUUGCGUUGAAUCCGAAUUUCAUAUUCAGGCCAGGACUC----- ((..(.(.(((.......(((..(--(((((........)))))))))(((((((.((...((((((..........))))))...))))))))))))).)..)).----- ( -26.20, z-score = 0.29, R) >droPer1.super_27 199140 104 - 1201981 GGGACAGAGCCGACUCCUUCUAAU--GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUUGGAUGUUGCGUUGAAUCCGAAUUUCAUAUUCAGGCCAGGACUC----- ((..(.(.(((.......(((..(--(((((........)))))))))(((((((.((...((((((..........))))))...))))))))))))).)..)).----- ( -26.20, z-score = 0.29, R) >droVir3.scaffold_13049 11345998 86 - 25233164 -------GCCUGCCUAAUGCCAACCAGUUGGACAGACAGUUGGCAAGAUGAAUGUCGACAGUUGGAUGAAA------AUCCAAAUUUCAUAUUCAGGCC------------ -------....((((..(((((((..(((.....))).)))))))....((((((.((...((((((....------))))))...)))))))))))).------------ ( -30.70, z-score = -4.02, R) >droMoj3.scaffold_6680 15275299 85 + 24764193 -------GCCAGCCUAAUGCCAACCAGUUGGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUG-AA------AUCCAAAUUUCAUAUUCAGGCC------------ -------(((..((...((((((....)))).))(((.(((((((.......))))))).)))))(((-((------((....))))))).....))).------------ ( -25.30, z-score = -2.51, R) >droGri2.scaffold_15110 7963440 86 - 24565398 -------GCCUGUCUAAUGCCAACCAGUUGGACGGACAGUUGGCAAGAUGAAUGUCGACAGUUGGAUGAAA------AUCCGAAUUUCAUAUUCAGGCC------------ -------((((((((..(((((((..(((.....))).)))))))))))((((((.((...((((((....------))))))...)))))))))))).------------ ( -31.80, z-score = -3.89, R) >consensus _______GCUCGACUC___CUAAU__GCCAGACAGACAGUUGGCAAGAUGAAUGUCGACAGUCGGAUGAAAU_CGGAAUCCGAAUUUCAUAUUCAGGCCAGGACUU_____ .........................................(((....(((((((.((...((((((..........))))))...))))))))).)))............ (-18.25 = -18.15 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:05 2011