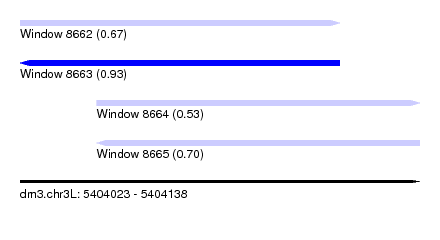
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,404,023 – 5,404,138 |
| Length | 115 |
| Max. P | 0.930662 |

| Location | 5,404,023 – 5,404,115 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.25473 |
| G+C content | 0.30228 |
| Mean single sequence MFE | -20.31 |
| Consensus MFE | -15.01 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672490 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 5404023 92 + 24543557 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- -------------.(((((((......--.))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).)))))---- ( -19.40, z-score = -1.83, R) >droPer1.super_27 188106 93 + 1201981 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUCG--- -------------.(((((((......--.))))))).((((.((......)).))))..(((((((((((((((.((.....)).....))))))))))).))))..--- ( -18.50, z-score = -1.49, R) >dp4.chrXR_group6 5065478 93 - 13314419 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUGG--- -------------.(((((((......--.))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).))))).--- ( -20.20, z-score = -1.82, R) >droEre2.scaffold_4784 8097280 92 + 25762168 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- -------------.(((((((......--.))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).)))))---- ( -19.40, z-score = -1.83, R) >droYak2.chr3L 5971289 92 + 24197627 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- -------------.(((((((......--.))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).)))))---- ( -19.40, z-score = -1.83, R) >droSec1.super_2 5349647 92 + 7591821 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUUGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- -------------.(((((((......--.))))))).((((.((......)).)))).(((((((((((((((((((.....)))....))))))))))).)))))---- ( -21.20, z-score = -2.10, R) >droSim1.chr3L 4920185 92 + 22553184 -------------UUAAGUUUUGUGCU--UGAACUUAUUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- -------------.(((((((......--.))))))).((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).)))))---- ( -19.40, z-score = -1.83, R) >droVir3.scaffold_13049 11334729 102 + 25233164 ---ACUAGAAUUGUGAACUUUUGUGUG--CGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUC---- ---.......((((((..(((((..((--((..((...((((.((......)).))))..)).))))..)))))))))))...((((((......))))))......---- ( -20.90, z-score = -1.64, R) >droMoj3.scaffold_6680 15264738 102 - 24764193 ---ACUAAAAUUGUGAACUUUUGUAUG--CGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- ---.......((((((..(((((..((--((..((...((((.((......)).))))..)).))))..)))))))))))...((((((......))))))......---- ( -20.40, z-score = -1.65, R) >droGri2.scaffold_15110 16552577 99 - 24565398 --------AAUUGCAAAGUUUUGUAUGUGCGAAAUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUUGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUC---- --------(((((((.((((((((....)))))))).......((......)))))))))((((((((((((((((((.....)))....))))))))))).)))).---- ( -22.20, z-score = -1.01, R) >droWil1.scaffold_180698 7190455 107 - 11422946 AUUCAUUCAUUGCCAAACCGCUUUAGG--UAAAGUU--UUUCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUGUGAA ........(((((..........((((--(((((((--.....))).)))))))).........))))).....(((((((..((((((......))))))..))))))). ( -22.01, z-score = -1.55, R) >droAna3.scaffold_13337 10154144 92 - 23293914 -------------UGAAGUUCUGUGCU--UGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG---- -------------..((((((......--.))))))..((((.((......)).)))).((((((((((((((((.((.....)).....))))))))))).)))))---- ( -20.70, z-score = -2.33, R) >consensus _____________UUAAGUUUUGUGCU__UGAACUUAUUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG____ ...............((((((.........))))))..((((.((......)).))))..(((((((((((((((.((.....)).....))))))))))).))))..... (-15.01 = -15.42 + 0.40)

| Location | 5,404,023 – 5,404,115 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.25473 |
| G+C content | 0.30228 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930662 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 5404023 92 - 24543557 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ----......(((((((((((.((.......))...)))))))))))......((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.70, z-score = -2.17, R) >droPer1.super_27 188106 93 - 1201981 ---CGAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ---.((....(((((((((((.((.......))...))))))))))).))...((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.20, z-score = -2.00, R) >dp4.chrXR_group6 5065478 93 + 13314419 ---CCAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ---.......(((((((((((.((.......))...)))))))))))......((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.00, z-score = -2.18, R) >droEre2.scaffold_4784 8097280 92 - 25762168 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ----......(((((((((((.((.......))...)))))))))))......((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.70, z-score = -2.17, R) >droYak2.chr3L 5971289 92 - 24197627 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ----......(((((((((((.((.......))...)))))))))))......((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.70, z-score = -2.17, R) >droSec1.super_2 5349647 92 - 7591821 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCAAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ----......(((((((((((...............)))))))))))......((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.06, z-score = -1.56, R) >droSim1.chr3L 4920185 92 - 22553184 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAAUAAGUUCA--AGCACAAAACUUAA------------- ----......(((((((((((.((.......))...)))))))))))......((((.(((....))).)))).((((((..--.......)))))).------------- ( -18.70, z-score = -2.17, R) >droVir3.scaffold_13049 11334729 102 - 25233164 ----GAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCG--CACACAAAAGUUCACAAUUCUAGU--- ----(((...(((((((((((.((.......))...)))))))))))......((((.(((....))).)))).....))).--........................--- ( -17.10, z-score = -1.26, R) >droMoj3.scaffold_6680 15264738 102 + 24764193 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCG--CAUACAAAAGUUCACAAUUUUAGU--- ----.........((((((((.....((.((((((((.((((.((((((......)).(((....))).)))).)))).)))--))...))).)).....))))))))--- ( -17.50, z-score = -1.11, R) >droGri2.scaffold_15110 16552577 99 + 24565398 ----GAAACAGCAAUUAAAAUAAAUUACAUUUUGCAAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAUUUCGCACAUACAAAACUUUGCAAUU-------- ----......(((((((((((...............)))))))))))....((((((((((....))).((.((....)).)).............)))))))-------- ( -18.26, z-score = -0.95, R) >droWil1.scaffold_180698 7190455 107 + 11422946 UUCACAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGAAA--AACUUUA--CCUAAAGCGGUUUGGCAAUGAAUGAAU ((((......(((((((((((.((.......))...)))))))))))(((.(((((((((((((.........--...))))--)))...)))))).)))..))))..... ( -24.10, z-score = -2.46, R) >droAna3.scaffold_13337 10154144 92 + 23293914 ----CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCA--AGCACAGAACUUCA------------- ----......(((((((((((.((.......))...)))))))))))......((((.(((....))).))))..((((((.--......))))))..------------- ( -20.10, z-score = -2.97, R) >consensus ____CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAAUAAGUUCA__AGCACAAAACUUAA_____________ ..........(((((((((((.((.......))...)))))))))))......((((.(((....))).))))...................................... (-15.70 = -15.78 + 0.08)
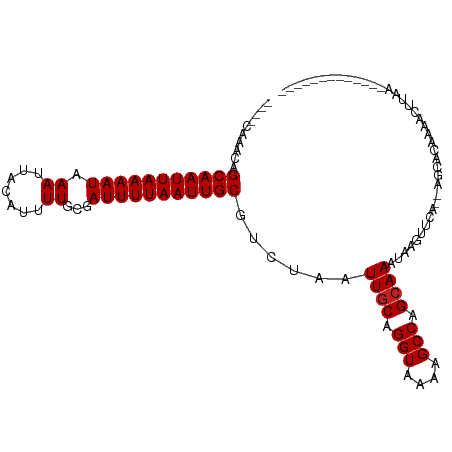
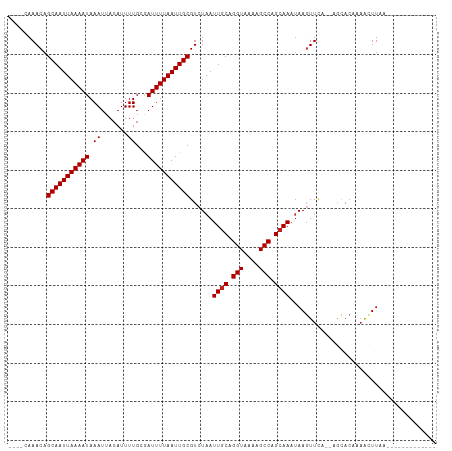
| Location | 5,404,045 – 5,404,138 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Shannon entropy | 0.18662 |
| G+C content | 0.32281 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529513 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 5404045 93 + 24543557 -----UUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAACUUUUUGGCCAAA-GCCU----------------- -----.((((.((......)).))))(((((((((((((((((.((.....)).....))))))))))).)))))-)...........(((....-))).----------------- ( -21.00, z-score = -1.82, R) >droPer1.super_27 188128 95 + 1201981 -----UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUCGAAACAACUUUUUGGCCAAAAGCCU----------------- -----......(((((((....((....(((((((((((((((.((.....)).....))))))))))).))))(((((.....))))))).))))))).----------------- ( -19.00, z-score = -1.16, R) >dp4.chrXR_group6 5065500 95 - 13314419 -----UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUGGAAACAACUUUUUGGCCAAAAGCCU----------------- -----......(((((((.((.......(((((((((((((((.((.....)).....))))))))))).))))(....)........))..))))))).----------------- ( -21.50, z-score = -1.59, R) >droEre2.scaffold_4784 8097302 93 + 25762168 -----UUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAACUUUUUGGCCAAA-GCCU----------------- -----.((((.((......)).))))(((((((((((((((((.((.....)).....))))))))))).)))))-)...........(((....-))).----------------- ( -21.00, z-score = -1.82, R) >droYak2.chr3L 5971311 93 + 24197627 -----UUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAACUUUUUGGCCAAA-GCCU----------------- -----.((((.((......)).))))(((((((((((((((((.((.....)).....))))))))))).)))))-)...........(((....-))).----------------- ( -21.00, z-score = -1.82, R) >droSec1.super_2 5349669 93 + 7591821 -----UUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUUGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAACUUUUUGGCCAAA-GCCU----------------- -----.((((.((......)).))))((((((((((((((((((((.....)))....))))))))))).)))))-)...........(((....-))).----------------- ( -22.80, z-score = -2.35, R) >droSim1.chr3L 4920207 93 + 22553184 -----UUUGCCGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAACUUUUUGGCCAAA-GCCU----------------- -----.((((.((......)).))))(((((((((((((((((.((.....)).....))))))))))).)))))-)...........(((....-))).----------------- ( -21.00, z-score = -1.82, R) >droVir3.scaffold_13049 11334761 92 + 25233164 -----UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUC-AAACAAGUU-UUGGCCAAA-GCCU----------------- -----...(((((((...((.((.....(((((((((((((((.((.....)).....))))))))))).)))).-...)).)).-..))))..)-))..----------------- ( -18.90, z-score = -0.84, R) >droMoj3.scaffold_6680 15264770 93 - 24764193 -----UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAAGUUUUUGGCCAAA-GCCU----------------- -----...(((((((...((.((...(((((((((((((((((.((.....)).....))))))))))).)))))-)..)).))....))))..)-))..----------------- ( -21.10, z-score = -1.38, R) >droGri2.scaffold_15110 16552606 93 - 24565398 -----UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUUGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUC-AAACAAGUUGUUGGCCAAA-GCAU----------------- -----..((((((((..((.((......((((((((((((((((((.....)))....))))))))))).)))).-.....)).))..))))..)-))).----------------- ( -22.90, z-score = -1.62, R) >droWil1.scaffold_180698 7190483 117 - 11422946 AAAGUUUUUCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUGUGAAGACAACGAAAAACAAAAAACAACAAUAACAACAAACCU ...(((((((....(((((..(((........)))..)))))(((((((..((((((......))))))..))))))).......)))))))......................... ( -21.10, z-score = -2.09, R) >droAna3.scaffold_13337 10154166 93 - 23293914 -----UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG-AAACAACUUUUUGGCCAAA-GCCU----------------- -----.((((.((......)).))))(((((((((((((((((.((.....)).....))))))))))).)))))-)...........(((....-))).----------------- ( -21.00, z-score = -1.80, R) >consensus _____UUUGCUGGCUUUUACCUGCAAUUAGACGCAAUUAAAAUCGCAAAAUGUAAUUUAUUUUAAUUGCUGUUUG_AAACAACUUUUUGGCCAAA_GCCU_________________ ......((((.((......)).))))..(((((((((((((((.((.....)).....))))))))))).))))..............(((.....))).................. (-16.32 = -16.73 + 0.42)
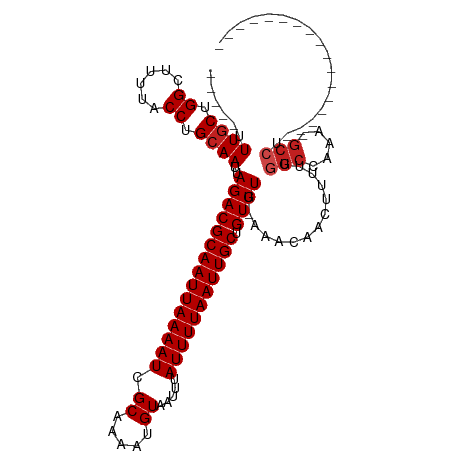
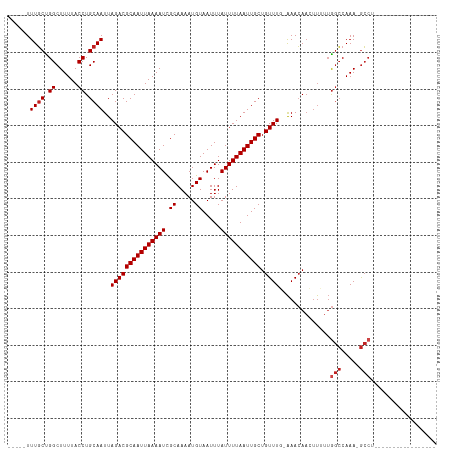
| Location | 5,404,045 – 5,404,138 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Shannon entropy | 0.18662 |
| G+C content | 0.32281 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -17.68 |
| Energy contribution | -17.94 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697274 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 5404045 93 - 24543557 -----------------AGGC-UUUGGCCAAAAAGUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAA----- -----------------.(((-(((.(((.....(((....-..))).(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -24.50, z-score = -2.28, R) >droPer1.super_27 188128 95 - 1201981 -----------------AGGCUUUUGGCCAAAAAGUUGUUUCGAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA----- -----------------.((((((((.((.....(((.......))).(((((((((((.((.......))...)))))))))))...........))))))))))......----- ( -23.80, z-score = -2.00, R) >dp4.chrXR_group6 5065500 95 + 13314419 -----------------AGGCUUUUGGCCAAAAAGUUGUUUCCAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA----- -----------------.((((((((.((.....(((.......))).(((((((((((.((.......))...)))))))))))...........))))))))))......----- ( -23.80, z-score = -2.07, R) >droEre2.scaffold_4784 8097302 93 - 25762168 -----------------AGGC-UUUGGCCAAAAAGUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAA----- -----------------.(((-(((.(((.....(((....-..))).(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -24.50, z-score = -2.28, R) >droYak2.chr3L 5971311 93 - 24197627 -----------------AGGC-UUUGGCCAAAAAGUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAA----- -----------------.(((-(((.(((.....(((....-..))).(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -24.50, z-score = -2.28, R) >droSec1.super_2 5349669 93 - 7591821 -----------------AGGC-UUUGGCCAAAAAGUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCAAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAA----- -----------------.(((-(((.(((.....(((....-..))).(((((((....(((......)))(((((....)))))...))))))).))).))))))......----- ( -24.10, z-score = -2.29, R) >droSim1.chr3L 4920207 93 - 22553184 -----------------AGGC-UUUGGCCAAAAAGUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCGGCAAA----- -----------------.(((-(((.(((.....(((....-..))).(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -24.50, z-score = -2.28, R) >droVir3.scaffold_13049 11334761 92 - 25233164 -----------------AGGC-UUUGGCCAA-AACUUGUUU-GAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA----- -----------------.(((-(((.(((..-....(((..-...)))(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -23.00, z-score = -1.76, R) >droMoj3.scaffold_6680 15264770 93 + 24764193 -----------------AGGC-UUUGGCCAAAAACUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA----- -----------------.(((-(((.(((.......(((..-...)))(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -23.00, z-score = -2.05, R) >droGri2.scaffold_15110 16552606 93 + 24565398 -----------------AUGC-UUUGGCCAACAACUUGUUU-GAAACAGCAAUUAAAAUAAAUUACAUUUUGCAAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA----- -----------------.(((-(..(((.....((((((..-......(((((((((((...............)))))))))))........))))))....)))))))..----- ( -22.65, z-score = -1.76, R) >droWil1.scaffold_180698 7190483 117 + 11422946 AGGUUUGUUGUUAUUGUUGUUUUUUGUUUUUCGUUGUCUUCACAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGAAAAACUUU .((((((((...((((((((((..((..............)).))))))))))...))))))))...(((((.(.(((((.(((((......))))).))))).))))))....... ( -21.64, z-score = -0.57, R) >droAna3.scaffold_13337 10154166 93 + 23293914 -----------------AGGC-UUUGGCCAAAAAGUUGUUU-CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA----- -----------------.(((-(((.(((.....(((....-..))).(((((((((((.((.......))...)))))))))))...........))).))))))......----- ( -24.50, z-score = -2.54, R) >consensus _________________AGGC_UUUGGCCAAAAAGUUGUUU_CAAACAGCAAUUAAAAUAAAUUACAUUUUGCGAUUUUAAUUGCGUCUAAUUGCAGGUAAAAGCCAGCAAA_____ ..................(((.....)))...................(((((((((((.((.......))...)))))))))))......((((.(((....))).))))...... (-17.68 = -17.94 + 0.26)

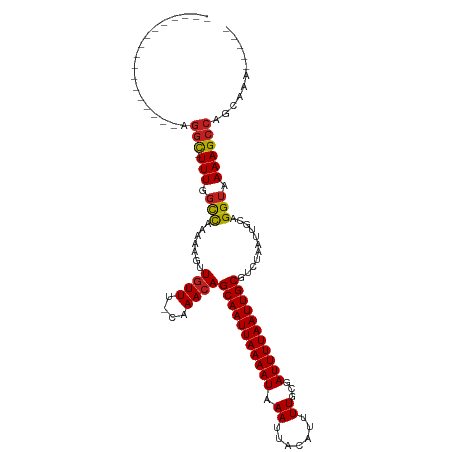
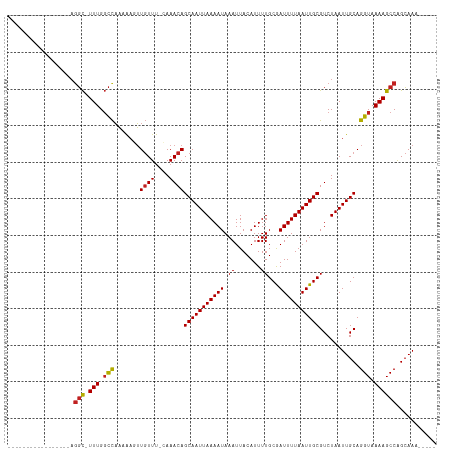
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:05:03 2011