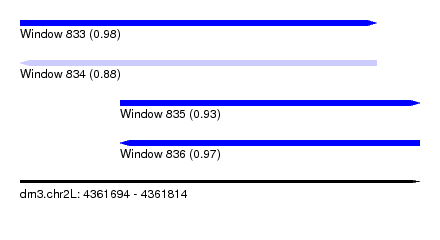
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,361,694 – 4,361,814 |
| Length | 120 |
| Max. P | 0.980007 |

| Location | 4,361,694 – 4,361,801 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.57962 |
| G+C content | 0.50638 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -17.43 |
| Energy contribution | -17.21 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
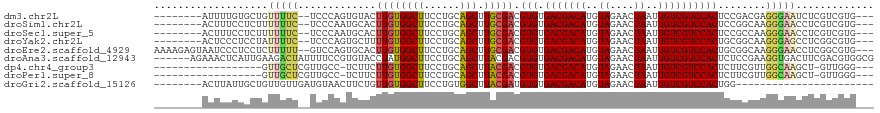
>dm3.chr2L 4361694 107 + 23011544 --------AUUUUGUGCUGUUUUC--UCCCAGUGUACUUGUGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGACGAGGGAAUCUCGUCGUG--- --------...(((((((((....--.(((((.....))).))......)))))..)))).(((.(((((((..((....))..))))))))))..(((((((.....)))))))..--- ( -33.40, z-score = -1.80, R) >droSim1.chr2L 4283595 107 + 22036055 --------ACUUUCCUCUUUUUUC--UCCCAAUGCACUUGUGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGGCAAGGGAACCUCGUCGUG--- --------................--........(((.((.((.(((((...(((.(.((.(((.(((((((..((....))..))))))))))))))))..))))))).))..)))--- ( -27.70, z-score = -0.87, R) >droSec1.super_5 2459055 107 + 5866729 --------ACUUUCCUCUUUUUUC--UCCCAAUGCACUUGUGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGCCAAGGGAACCUCGUCGUG--- --------................--((((...((...((..(....)..))))..(((..(((.(((((((..((....))..))))))))))..)))...))))...........--- ( -26.90, z-score = -1.18, R) >droYak2.chr2L 4395115 107 + 22324452 --------ACUCCCUCCUAUUUUC--UCCCAGUGCUUUUGUGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUGCGGCAAGGGAGCCUCGGCGUG--- --------................--.......(((...(.((((((((...(((.(((..(((.(((((((..((....))..)))))))))))))))).)))))))).))))...--- ( -35.30, z-score = -1.60, R) >droEre2.scaffold_4929 4443831 115 + 26641161 AAAAGAGUAAUCCCUCCUCUUUUU--GUCCAGUGCACUUGUGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUGCGGCAAGGGAACCUCGGCGUG--- ....(((...(((((........(--(((((((((((((((((((......))).))))).))).(((((((..((....))..)))))))))))).)))))))))..)))......--- ( -35.00, z-score = -0.92, R) >droAna3.scaffold_12943 1538018 114 - 5039921 ------AGAAACUCAUUGAAGACUAUUUUCCGUGUACCUAUGGCUUCCUGCAGCUUACGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCUCCGAAGGUGACUUCGACGUGGCG ------...........((((.((((.............)))))))).....(((.(((..(((.(((((((..((....))..))))))))))....(((((....))))).)))))). ( -32.92, z-score = -1.60, R) >dp4.chr4_group3 2285085 97 - 11692001 ------------------GUUGCUCGUUGCC-UCUUCUUGUGGCUUCCUGCAGCUUACGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCUUCGUUGGCAAGCU-GUUGGG--- ------------------..........(((-.(.....).)))..((.(((((((.(((((.(((((((((..((....))..)))))).)))....)))))..)))))-)).)).--- ( -34.20, z-score = -2.98, R) >droPer1.super_8 3370647 97 - 3966273 ------------------GUUGCUCGUUGCC-UCUUCUUGUGGCUUCCUGCAGCUUACGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCUUCGUUGGCAAGCU-GUUGGG--- ------------------..........(((-.(.....).)))..((.(((((((.(((((.(((((((((..((....))..)))))).)))....)))))..)))))-)).)).--- ( -34.20, z-score = -2.98, R) >droGri2.scaffold_15126 4476195 89 - 8399593 --------ACUUAUUGCUGUUGUUGAUGUAACUUCUGUUGUGGCUUCCUGUGGCUUACGAUGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUGG----------------------- --------..........((((......))))....(((((((((......))).))))))(((.(((((((..((....))..))))))))))...----------------------- ( -20.60, z-score = -0.98, R) >consensus ________ACUUUCUUCUGUUUUU__UCCCAGUGCACUUGUGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGCCAAGGGAACCUCGUCGUG___ ...................(((((.............((((((((......))).))))).(((.(((((((..((....))..))))))))))........)))))............. (-17.43 = -17.21 + -0.22)
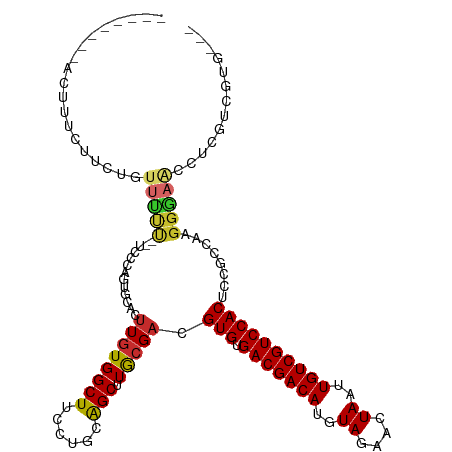
| Location | 4,361,694 – 4,361,801 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.57962 |
| G+C content | 0.50638 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.59 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4361694 107 - 23011544 ---CACGACGAGAUUCCCUCGUCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCACAAGUACACUGGGA--GAAAACAGCACAAAAU-------- ---..(((((((.....)))))))((((((((((((..((....))..))))))).))).))....((((.......((...((....)).)).--.....)))).......-------- ( -33.12, z-score = -2.53, R) >droSim1.chr2L 4283595 107 - 22036055 ---CACGACGAGGUUCCCUUGCCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCACAAGUGCAUUGGGA--GAAAAAAGAGGAAAGU-------- ---..........(((((.(((((..((((((((((..((....))..))))))).)))..))...(((......))).......)))..))))--)...............-------- ( -28.00, z-score = -0.04, R) >droSec1.super_5 2459055 107 - 5866729 ---CACGACGAGGUUCCCUUGGCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCACAAGUGCAUUGGGA--GAAAAAAGAGGAAAGU-------- ---..........(((((.(((((..((((((((((..((....))..))))))).)))..)))..(((......)))........))..))))--)...............-------- ( -29.70, z-score = -0.59, R) >droYak2.chr2L 4395115 107 - 22324452 ---CACGCCGAGGCUCCCUUGCCGCAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCACAAAAGCACUGGGA--GAAAAUAGGAGGGAGU-------- ---..(.((....(((((.(((.((.((((((((((..((....))..))))))).)))...))..(((......))).......)))..))))--)......)).).....-------- ( -32.40, z-score = -0.55, R) >droEre2.scaffold_4929 4443831 115 - 26641161 ---CACGCCGAGGUUCCCUUGCCGCAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCACAAGUGCACUGGAC--AAAAAGAGGAGGGAUUACUCUUUU ---(((...(.(((((((((((.((.((((((((((..((....))..))))))).))))).))))).....)).)))).)..)))........--....(((((((......))))))) ( -33.10, z-score = -0.42, R) >droAna3.scaffold_12943 1538018 114 + 5039921 CGCCACGUCGAAGUCACCUUCGGAGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGUAAGCUGCAGGAAGCCAUAGGUACACGGAAAAUAGUCUUCAAUGAGUUUCU------ .((.(((((((((....))))))...((((((((((..((....))..))))))).)))..)))..))....(((((((((.((...((........))..)).))).))))))------ ( -28.10, z-score = -0.04, R) >dp4.chr4_group3 2285085 97 + 11692001 ---CCCAAC-AGCUUGCCAACGAAGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGUAAGCUGCAGGAAGCCACAAGAAGA-GGCAACGAGCAAC------------------ ---.((..(-(((((....((((...((((((((((..((....))..))))))).))).))))))))))..))..(((.(.....).-)))..........------------------ ( -30.80, z-score = -3.12, R) >droPer1.super_8 3370647 97 + 3966273 ---CCCAAC-AGCUUGCCAACGAAGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGUAAGCUGCAGGAAGCCACAAGAAGA-GGCAACGAGCAAC------------------ ---.((..(-(((((....((((...((((((((((..((....))..))))))).))).))))))))))..))..(((.(.....).-)))..........------------------ ( -30.80, z-score = -3.12, R) >droGri2.scaffold_15126 4476195 89 + 8399593 -----------------------CCAGUGGACGACAAUUAGUUCUACAUGUCGUCACACAUCGUAAGCCACAGGAAGCCACAACAGAAGUUACAUCAACAACAGCAAUAAGU-------- -----------------------...((((((((((..((....))..))))))).))).......((....((...)).........(((.....)))....)).......-------- ( -14.10, z-score = -0.85, R) >consensus ___CACGACGAGGUUCCCUUGGCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCACAAGUGCACUGGAA__AAAAACAGAAGAAAGU________ ..............(((.........((((((((((..((....))..))))))).))).......(((......)))............)))........................... (-15.32 = -15.59 + 0.27)

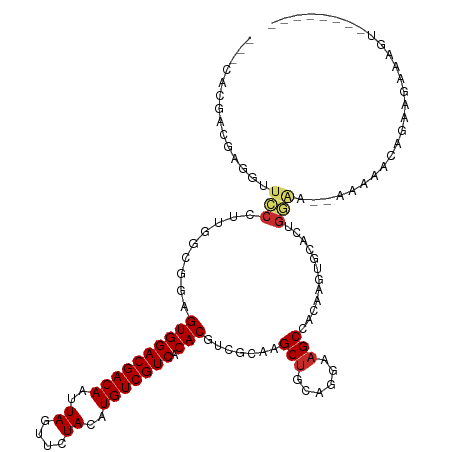
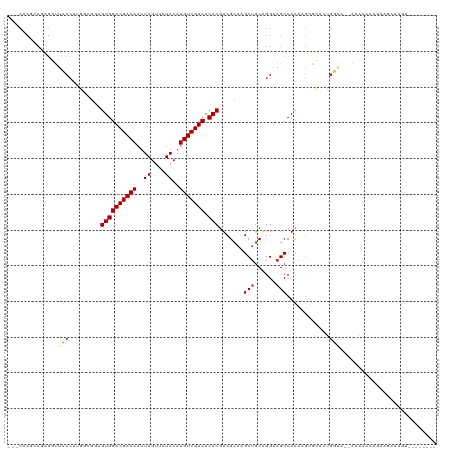
| Location | 4,361,724 – 4,361,814 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.01 |
| Shannon entropy | 0.46310 |
| G+C content | 0.56297 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -16.81 |
| Energy contribution | -18.56 |
| Covariance contribution | 1.74 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4361724 90 + 23011544 UGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGACGAGGGAAUCUCGUCGUGCAGGAGGAGUCGG--------------------------- ...(((((((((......((.(((.(((((((..((....))..))))))))))))(((((((.....))))))))))))))))......--------------------------- ( -39.70, z-score = -3.16, R) >droSim1.chr2L 4283625 90 + 22036055 UGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGGCAAGGGAACCUCGUCGUGCUGGAGGAGCCGG--------------------------- .((((.(((.((((..((((((((.(((((((..((....))..))))))))))(((......))).....))))))))).)))))))..--------------------------- ( -42.70, z-score = -3.92, R) >droSec1.super_5 2459085 90 + 5866729 UGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGCCAAGGGAACCUCGUCGUGCAGGAGGAGCCGG--------------------------- .((((((((((.....((((((((.(((((((..((....))..))))))))))(((......))).....)))))))))))..))))..--------------------------- ( -38.40, z-score = -2.80, R) >droYak2.chr2L 4395145 90 + 22324452 UGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUGCGGCAAGGGAGCCUCGGCGUGCAGGAGGAGUUGG--------------------------- ...((((((((((((.(((..(((.(((((((..((....))..)))))))))))))(((......)))..))).)))))))))......--------------------------- ( -38.20, z-score = -2.21, R) >droEre2.scaffold_4929 4443869 90 + 26641161 UGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUGCGGCAAGGGAACCUCGGCGUGCAGGAGGAGCUGG--------------------------- .(((((((((((.....((..(((.(((((((..((....))..))))))))))..))((..((....))..)).)))))))..))))..--------------------------- ( -35.00, z-score = -1.38, R) >droAna3.scaffold_12943 1538052 117 - 5039921 UGGCUUCCUGCAGCUUACGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCUCCGAAGGUGACUUCGACGUGGCGUGGCGGAUAGGGGCCUUCUGCAGAGAAGGGGAGCUGG .((((((((((.(((.(((..(((.(((((((..((....))..))))))))))....(((((....))))).))))))...)))).......((((((....)))))))))))).. ( -49.20, z-score = -2.66, R) >dp4.chr4_group3 2285106 92 - 11692001 UGGCUUCCUGCAGCUUACGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCUUCGUUGGCAAGCU-GUUGGGUAGGGGGACUCAGGCU------------------------ .(((((((.(((((((.(((((.(((((((((..((....))..)))))).)))....)))))..)))))-)).))...((....)).)))))------------------------ ( -34.60, z-score = -2.04, R) >droPer1.super_8 3370668 92 - 3966273 UGGCUUCCUGCAGCUUACGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCUUCGUUGGCAAGCU-GUUGGGUAGGGGGACUCAGGCU------------------------ .(((((((.(((((((.(((((.(((((((((..((....))..)))))).)))....)))))..)))))-)).))...((....)).)))))------------------------ ( -34.60, z-score = -2.04, R) >droWil1.scaffold_180772 4638279 75 - 8906247 UGGCUUCCUGCAGCUUACGACGUGU----------AGAACUAAUUGUUGUCCAA-GAGAGGAGAGGAGCAGGAAAGGGAAUGCCAC------------------------------- (((((((((((..(((.(((((..(----------(....))..)))))(((..-....))))))..))))))).......)))).------------------------------- ( -20.61, z-score = -1.05, R) >consensus UGGCUUCCUGCAGCUUGCGACGUGUGACGACAUGUAGAACUAAUUGUCGUCCACUCCGCCAAGGGAACCUCGUCGUGCAGGAGGAGUCGG___________________________ ...((((((((.(....)...(((.(((((((..((....))..))))))))))......................))))))))................................. (-16.81 = -18.56 + 1.74)

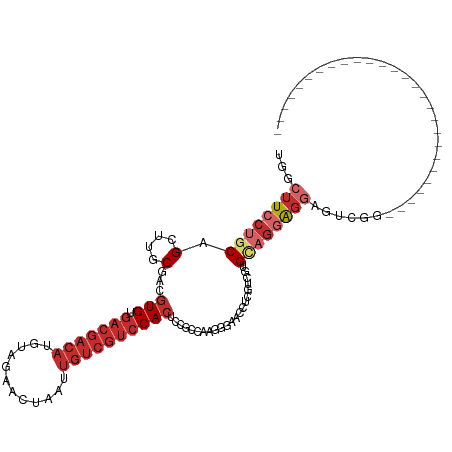
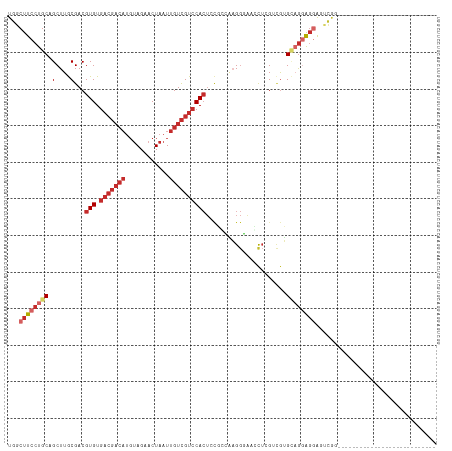
| Location | 4,361,724 – 4,361,814 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.01 |
| Shannon entropy | 0.46310 |
| G+C content | 0.56297 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -12.18 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4361724 90 - 23011544 ---------------------------CCGACUCCUCCUGCACGACGAGAUUCCCUCGUCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCA ---------------------------........((((((((((((((.....)))))))((((((((((((..((....))..))))))).))).))......)))))))..... ( -36.60, z-score = -3.65, R) >droSim1.chr2L 4283625 90 - 22036055 ---------------------------CCGGCUCCUCCAGCACGACGAGGUUCCCUUGCCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCA ---------------------------..(((((((.((((.((((..(((......)))...((((((((((..((....))..))))))).)))))))...)))).))).)))). ( -39.30, z-score = -3.95, R) >droSec1.super_5 2459085 90 - 5866729 ---------------------------CCGGCUCCUCCUGCACGACGAGGUUCCCUUGGCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCA ---------------------------..((((..((((((((..(((((...)))))(((..((((((((((..((....))..))))))).)))..)))..).))))))))))). ( -42.40, z-score = -4.34, R) >droYak2.chr2L 4395145 90 - 22324452 ---------------------------CCAACUCCUCCUGCACGCCGAGGCUCCCUUGCCGCAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCA ---------------------------........(((((((.((...(((......)))((.((((((((((..((....))..))))))).)))...))..)))))))))..... ( -34.20, z-score = -3.26, R) >droEre2.scaffold_4929 4443869 90 - 26641161 ---------------------------CCAGCUCCUCCUGCACGCCGAGGUUCCCUUGCCGCAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCA ---------------------------...(((..(((((((.(((((((...)))))..((.((((((((((..((....))..))))))).)))...))..)))))))))))).. ( -34.40, z-score = -3.04, R) >droAna3.scaffold_12943 1538052 117 + 5039921 CCAGCUCCCCUUCUCUGCAGAAGGCCCCUAUCCGCCACGCCACGUCGAAGUCACCUUCGGAGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGUAAGCUGCAGGAAGCCA .........((((.((((((..(((........)))(((.(.(..(((((....)))))..).((((((((((..((....))..))))))).)))).)))...))))))))))... ( -39.40, z-score = -2.75, R) >dp4.chr4_group3 2285106 92 + 11692001 ------------------------AGCCUGAGUCCCCCUACCCAAC-AGCUUGCCAACGAAGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGUAAGCUGCAGGAAGCCA ------------------------..((((((.....))......(-(((((....((((...((((((((((..((....))..))))))).))).))))))))))))))...... ( -27.60, z-score = -2.33, R) >droPer1.super_8 3370668 92 + 3966273 ------------------------AGCCUGAGUCCCCCUACCCAAC-AGCUUGCCAACGAAGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGUAAGCUGCAGGAAGCCA ------------------------..((((((.....))......(-(((((....((((...((((((((((..((....))..))))))).))).))))))))))))))...... ( -27.60, z-score = -2.33, R) >droWil1.scaffold_180772 4638279 75 + 8906247 -------------------------------GUGGCAUUCCCUUUCCUGCUCCUCUCCUCUC-UUGGACAACAAUUAGUUCU----------ACACGUCGUAAGCUGCAGGAAGCCA -------------------------------.((((.......(((((((.....(((....-..)))........((((.(----------((.....)))))))))))))))))) ( -18.71, z-score = -1.83, R) >consensus ___________________________CCGACUCCUCCUGCACGACGAGGUUCCCUUGGCGGAGUGGACGACAAUUAGUUCUACAUGUCGUCACACGUCGCAAGCUGCAGGAAGCCA ...............................................................((((((((((............))))))).))).......(((......))).. (-12.18 = -13.07 + 0.89)

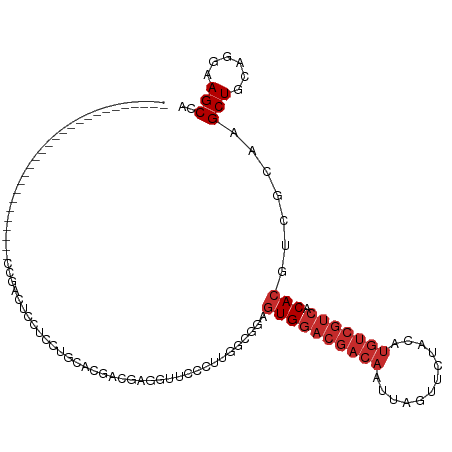
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:17 2011