
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 490,028 – 490,120 |
| Length | 92 |
| Max. P | 0.618180 |

| Location | 490,028 – 490,120 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 67.97 |
| Shannon entropy | 0.63410 |
| G+C content | 0.36893 |
| Mean single sequence MFE | -18.56 |
| Consensus MFE | -4.03 |
| Energy contribution | -4.08 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
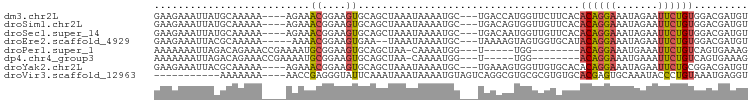
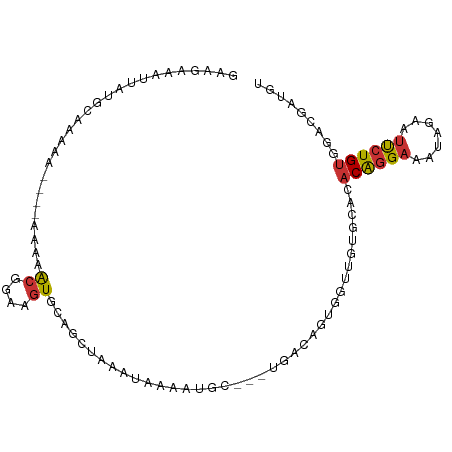
>dm3.chr2L 490028 92 + 23011544 GAAGAAAUUAUGCAAAAA----AGAAACGGAAGUGCAGCUAAAUAAAAUGC---UGACCAUGGUUCUUCACACAGGAAAUAGAAUUCUGUGGACGAUGU ((((((.(((((......----....((....)).((((..........))---))..))))))))))).(((((((.......)))))))........ ( -23.00, z-score = -3.23, R) >droSim1.chr2L 496521 92 + 22036055 GAAGAAAUUAUGCAAAAA----AGAAACGGAAGUGCAGCUAAAUAAAAUGC---UGACAGUGGUUGUUCACACAGGAAAUAGAAUUCUGUGGACGAUGU (((..((((((.......----....((....)).((((..........))---))...)))))).))).(((((((.......)))))))........ ( -18.00, z-score = -1.47, R) >droSec1.super_14 473041 92 + 2068291 GAAGAAAUUAUGCAAAAA----AGAAACGGAAGUGCAGCUAAAUAAAAUGC---UGACAAUGGUUGUUCACACAGGAAAUAGAAUUCUGUGGACGAUGU (((..((((((.......----....((....)).((((..........))---))...)))))).))).(((((((.......)))))))........ ( -17.70, z-score = -1.75, R) >droEre2.scaffold_4929 542474 89 + 26641161 GAAGAAAUUACGCAAAAA-----AAAACGGAAGUGAA--UAAAUAAAAUGC---UAAAAGUAGUGGUGCAUACAGGAAAUAGAAUUCUGUGGACGAUGU ...........(((....-----...((....))...--..........((---((....))))..))).(((((((.......)))))))........ ( -11.90, z-score = -1.41, R) >droPer1.super_1 7676125 82 - 10282868 AAAAAAAUUAGACAGAAACCGAAAAUGCGGAAGUGCAGCUAA-CAAAAUGG---U-----UGG--------ACAGGAAAUGAAAUUCUGUCAGUGAAAG ..........(((((((.(((......)))..((.((((((.-.....)))---)-----)).--------))...........)))))))........ ( -21.50, z-score = -4.50, R) >dp4.chr4_group3 10555911 82 - 11692001 AAAAAAAUUAGACAGAAACCGAAAAUGCGGAAGUGCAGCUAA-CAAAAUGG---U-----UGG--------ACAGGAAAUGAAAUUCUGUCAGUGAAAG ..........(((((((.(((......)))..((.((((((.-.....)))---)-----)).--------))...........)))))))........ ( -21.50, z-score = -4.50, R) >droYak2.chr2L 474666 92 + 22324452 GAAGAAAUUACGCAAAAA----AGAAACGGAAGUGCAGCUAAAUAAAAUGC---UGAAAGUGGUUGUGCACACAGGAAAUAGAAUUCUGCGGACGAUGU ..........(((.....----....((....)).((((..........))---))...)))(((((.(.(.(((((.......))))).))))))).. ( -16.50, z-score = -0.84, R) >droVir3.scaffold_12963 18913068 84 + 20206255 -----------AAAAAAA----AACCGAGGGUAUUCAAAUAAAUAAAAUGUAGUCAGGCGUGCGCGUGUGCACGAGUGCAAAUACCCUGUAAAUGAGGU -----------.......----.....((((((((.............((((.((....(((((....))))))).))))))))))))........... ( -18.41, z-score = -1.20, R) >consensus GAAGAAAUUAUGCAAAAA____AAAAACGGAAGUGCAGCUAAAUAAAAUGC___UGACAGUGGUUGUGCACACAGGAAAUAGAAUUCUGUGGACGAUGU ..........................((....)).....................................((((((.......))))))......... ( -4.03 = -4.08 + 0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:57 2011