| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,354,575 – 5,354,737 |
| Length | 162 |
| Max. P | 0.688157 |

| Location | 5,354,575 – 5,354,737 |
|---|---|
| Length | 162 |
| Sequences | 15 |
| Columns | 170 |
| Reading direction | reverse |
| Mean pairwise identity | 74.74 |
| Shannon entropy | 0.58419 |
| G+C content | 0.55519 |
| Mean single sequence MFE | -55.43 |
| Consensus MFE | -29.21 |
| Energy contribution | -29.25 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
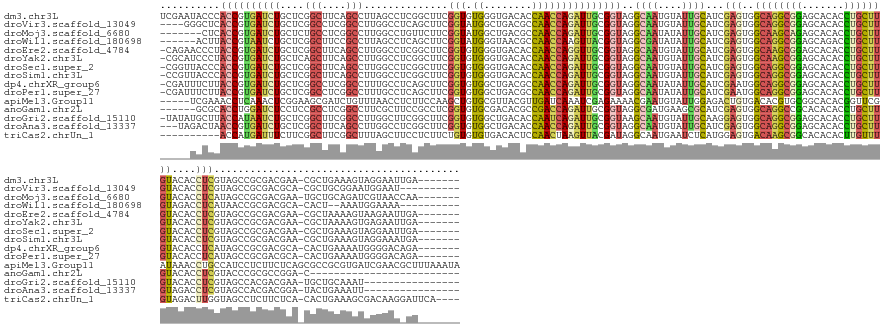
>dm3.chr3L 5354575 162 - 24543557 UCGAAUACCCACCGUGAUCUGCUCGGCUUCAGCCUUAGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGAA-CGCUGAAAGUAGGAAUUGA------- .......((.((((..(((((...(((....)))..(((....)))..(((((.....)))))...)))))..)))).))((((...((((..((((((((((((((.......)))))))).)).)))).....(((.....-))).....))))..)))).------- ( -55.80, z-score = 0.01, R) >droVir3.scaffold_13049 8125529 155 - 25233164 ----GGGCUCACCGUGAUCUGCUCGGCCUCGGCCUUGGCCUCAGCUUCGGUAUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGCA-CGCUGCGGAAUGGAAU---------- ----..((..((((..(((((...(((....)))(((((.((((((.......)))))).))))).)))))..))))..))..(.((((.(..((((((((((((((.......)))))))).)).))))..((.(((.....-))).))).)))).)..---------- ( -66.80, z-score = -1.37, R) >droMoj3.scaffold_6680 11788428 155 - 24764193 -------CUCACCGUGAUCUGCUCUGCCUCGGCCUUGGCCUGUUCUUCGGUAUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAGUGGCAAGCAGAGCACACCUGCUUGUACACCUCAUAGCCGCGACGAA-UGCUGCAGAUCGUAACCAA------- -------........(((((((...((...(((....))).((((.(((((.((((....))))))).....(((((..((((....))))...(((((((((((((.......)))))))).)).)))...))))))).)))-))).)))))))........------- ( -59.50, z-score = -2.69, R) >droWil1.scaffold_180698 7120483 151 + 11422946 ------ACUUACCGUAAUCUGCUCGGCUUCCGCCUUAGCCUCAGCUUCGGUAUGGGUAACGCCAACCAAGUUACGGUAGGCGAUAUAUUGCAUCGAGUGGCAGGCGGAGCAGACCUGCUUGUAGACCUCAUAACCGCGACGCA-CACU--AAAUGGAAAA---------- ------.....((((..(((((((.(((((((((...((((..((....))..))))..((((.(((.......))).))))............).)))).)))).)))))))..((((((..(.........)..))).)))-....--..))))....---------- ( -49.00, z-score = -0.98, R) >droEre2.scaffold_4784 8045937 161 - 25762168 -CAGAACCCUACCGUGAUCUGCUCGGCUUCAGCCUUGGCCUCGGCUUCGGUGUGGGUGACACCAACCAGGUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAAGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGAA-CGCUAAAAGUAAGAAUUGA------- -......(((((((..(((((...(((....)))..(((....)))..(((((.....)))))...)))))..)))))))((((...((((..((((((((((((((.......)))))))).)).)))).....(((.....-))).....))))..)))).------- ( -60.10, z-score = -1.21, R) >droYak2.chr3L 5920842 161 - 24197627 -CGCAUCCCUACCGUGAUCUGCUCAGCUUCAGCCUUGGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAAGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGAA-CGCUAAAAGUGAGAAUUGA------- -(((...(((((((..(((((...((((...((....))...))))..(((((.....)))))...)))))..))))))).........((..((((((((((((((.......)))))))).)).))))..)).))).....-(((.....)))........------- ( -59.20, z-score = -1.19, R) >droSec1.super_2 5300244 161 - 7591821 -CGGUUACCCACCGUGAUCUGCUCGGCUUCAGCCUUGGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGAA-CGCUGAAAGUAGGAAUUGA------- -((((.....))))...((((((.((((........))))(((((((((((.(((......))))))...(((((((..((((....))))..((((((((((((((.......)))))))).)).))))..))))))).)))-.))))).))))))......------- ( -58.20, z-score = 0.11, R) >droSim1.chr3L 4869602 161 - 22553184 -CCGUUACCCACCGUGAUCUGCUCGGCUUCAGCCUUGGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGAA-CGCUGAAAGUAGGAAAUGA------- -(((((((((((..(((...(((.((((........))))..))).)))..)))))))))......(((.(((((((..((((....))))..((((((((((((((.......)))))))).)).))))..)))))))....-..)))......))......------- ( -57.80, z-score = -0.01, R) >dp4.chrXR_group6 5006111 161 + 13314419 -CGAUUUCUUACCGUGAUCUGCUCGGCCUCGGCCUUUGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAAUGGCAGGCGGAGCACACCUGCUUGUACACCUCAUAGCCGCGACGCA-CACUGAAAAUGGGGACAGA------- -..(((.(((((((..(((((...(((.((((((..((((........)))).)))))).)))...)))))..))))))).)))....(((.(((...((((((.((((((....)))))...).)))....))).))).)))-.........((....))..------- ( -59.00, z-score = -1.48, R) >droPer1.super_27 128815 161 - 1201981 -CGAUUUCUUACCGUGAUCUGCUCGGCCUCGGCCUUUGCCUCAGCUUCGGUGUGGCUGACGCCAACCAGAUUGCGGUAGGCAAUAUAUUGCAUCGAAUGGCAGGCGGAGCACACCUGCUUGUACACCUCAUAGCCGCGACGCA-CACUGAAAAUGGGGACAGA------- -..(((.(((((((..(((((...(((.((((((..((((........)))).)))))).)))...)))))..))))))).)))....(((.(((...((((((.((((((....)))))...).)))....))).))).)))-.........((....))..------- ( -59.00, z-score = -1.48, R) >apiMel3.Group11 133081 165 - 12576330 -----UCGAAACCUCAAACUCGGAAGCGAUCUGUUUAACCUCUUCCAAGCUGUGCGUUACGUUGAUCAAAUCGAGAAAACGAAUGUAUUGGAGACUGUGACACGUGCGGCACACGGUUCGAUAAACCUGCCAUCCUCUUCUCAGCGCCGCGUGAUCGAACGCUUUAAAUA -----.(((..........)))((((((.((((((.....(((.(((....(((((((.((((..((.....))...)))))))))))))))))....))))((((((((....((((.....)))).((.............))))))))))...)).))))))..... ( -39.42, z-score = 0.54, R) >anoGam1.chr2L 39590018 138 - 48795086 ------GCGCACCUGGAUCUCCUCCGCCUCGGCCUUCGCUUCCGCCUCGGUGUGCGACACGCCGACCAGAUUGCGGUAGGCGAUGAAGCGCAUCGAGUGGCAGGCCGCACACACCUGCUUGUACACCUCGUACCCGCGCCGGA-C------------------------- ------(((.....((....))..))).(((((....((((((((.(((((((((..((((((.(((.......))).)))).))..))))))))))))).))))...........((..((((.....))))..))))))).-.------------------------- ( -54.40, z-score = -0.62, R) >droGri2.scaffold_15110 7154473 152 - 24565398 -UAUAUGCUUACCAUAAUCUGCUCGGCUUCGGCCUUGGCUUCGGCUUCGGUGUGGCUGACACCAAUCAGAUUGCGGUAAGCAAUGUAUUGCAAGGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUAGCCACGACGAA-UGCUGCAAAU---------------- -....((((((((.(((((((...(((....)))((((..((((((.......))))))..)))).))))))).)))))))).((((..(((.(..(((((((((((.......)))))).(((.....))))))))..)...-)))))))...---------------- ( -60.60, z-score = -2.88, R) >droAna3.scaffold_13337 10101366 150 + 23293914 ---UAGACUAACCGUGAUCUGCUCGGCUUCAGCCUUGGCCUCGGCUUCGGUGUGGCUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUAGACCUCGUAGCCACGACGGA-UACUGAAAUU---------------- ---....(((.(((..(((((...((..((((((...(((........)))..))))))..))...)))))..))))))((((....)))).((.((((((((((((.......))))))))...((((((....)))).)).-))))))....---------------- ( -53.90, z-score = -0.95, R) >triCas2.chrUn_1 388099 155 + 1280983 ----------ACCAUGAUUUCUUCGGCUUCGGCUUUAGCUUCCUCUUCUGUGUGUGACACUCCAACUAAGUUACGAUAGGCAAUGAAUCUCAUGGAGUGACAAGCGGCACACACUUGUUUGUAGACUUGGUAGCCUCUUCUCA-CACUGAAAGCGACAAGGAUUCA---- ----------....(((.(((((.(((....)))...((((.....((.(((((.((((((((......(((......))).(((.....))))))))).(((((.(((.((....)).))).).)))).........)).))-))).))))))...))))).)))---- ( -38.70, z-score = 0.32, R) >consensus _____UACUCACCGUGAUCUGCUCGGCUUCGGCCUUGGCCUCGGCUUCGGUGUGGGUGACACCAACCAGAUUGCGGUAGGCAAUGUAUUGCAUCGAGUGGCAGGCGGAGCACACCUGCUUGUACACCUCGUAGCCGCGACGAA_CGCUGAAAGUGGGAAU_GA_______ ..........((((((((((....(((....)))..............(((.((........)))))))))))))))..((((....))))...(((..((((((((.......))))))))....)))......................................... (-29.21 = -29.25 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:56 2011