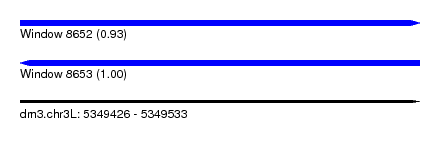
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,349,426 – 5,349,533 |
| Length | 107 |
| Max. P | 0.995863 |

| Location | 5,349,426 – 5,349,533 |
|---|---|
| Length | 107 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Shannon entropy | 0.37224 |
| G+C content | 0.48850 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.53 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
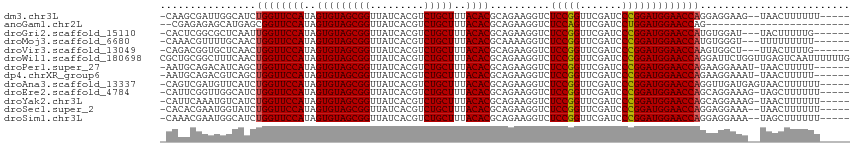
>dm3.chr3L 5349426 107 + 24543557 -CAAGCGAUUGGCAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGGAGGAAG--UAACUUUUUU----- -.(((..(((..(..(((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))..)..))--)..)))....----- ( -33.50, z-score = -1.46, R) >anoGam1.chr2L 45432148 89 + 48795086 --CGAGAGAGCAUGAGCGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCAGUUCGAUCCUGGAUGGAACCAG------------------------ --.......((....))((((((((.(((((((((.........)))))..)))).(((..((((........))))))).))))))))..------------------------ ( -27.20, z-score = -1.01, R) >droGri2.scaffold_15110 7150282 105 + 24565398 -CACUCGGCGCUCAAUUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAUGUGGAU---UACUUUUUG------ -.......(((.....((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))).)))...---.........------ ( -29.60, z-score = -0.66, R) >droMoj3.scaffold_6680 11783997 105 + 24764193 -CAAACGUUUUGCAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAAAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAUGUGGGU---UUUUUUUUU------ -.......((..((..((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))))..)).---.........------ ( -30.50, z-score = -1.62, R) >droVir3.scaffold_13049 8121241 105 + 25233164 -CAGACGGUGCUCAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAAGUGGCU---UUACUUUUG------ -((((.(..((.((..((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))..)))).---...).))))------ ( -32.80, z-score = -1.22, R) >droWil1.scaffold_180698 7114872 115 - 11422946 CGCUGCGGCUUUCAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGGAUUCUGGUUGAGUCAAUUUUUUG .((....))......(((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))(((((.....)))))......... ( -37.40, z-score = -1.75, R) >droPer1.super_27 123157 107 + 1201981 -AAUGCAGACAUCAGCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGAAGGAAAU-UAACUUUUU------ -..........((..(((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))...))...-.........------ ( -32.50, z-score = -1.97, R) >dp4.chrXR_group6 5000531 107 - 13314419 -AAUGCAGACGUCAGCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGAAGGAAAU-UAACUUUUU------ -..........((..(((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))...))...-.........------ ( -32.40, z-score = -1.65, R) >droAna3.scaffold_13337 10096500 109 - 23293914 -CAGUCGAUGUUCAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGGUUGAUGAGUAACUUUUUU----- -.(((......(((((((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))).)))......))).....----- ( -34.00, z-score = -1.79, R) >droEre2.scaffold_4784 8040760 108 + 25762168 -CAUUCGGUUGGCAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGCAGGAAAG-UAGCUUUUUU----- -.....(((((.(..(((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))...)....-))))).....----- ( -31.60, z-score = -0.92, R) >droYak2.chr3L 5915726 108 + 24197627 -CAUUCAAAUGUCAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGCAGGAAAG-UAACUUUUUU----- -..........((..(((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))))...))...-..........----- ( -31.30, z-score = -2.04, R) >droSec1.super_2 5294685 107 + 7591821 -CACACGAAUGGUAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGGAGGAAA--UAACUUUUUU----- -.((((..((((.........)))).))))...((((((..(.(((((.......))))).).((((((((((.(((...))).))))).)))))..)--))))).....----- ( -31.80, z-score = -1.38, R) >droSim1.chr3L 4864024 107 + 22553184 -CAAACGAAUGGCAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGGAGGAAA--UAGCUUUUUU----- -.............((((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))))......--..........----- ( -31.70, z-score = -1.14, R) >consensus _CAUACGGAUGUCAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAGGAGGAAA__UAACUUUUUU_____ ................((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))......................... (-27.60 = -27.53 + -0.06)
| Location | 5,349,426 – 5,349,533 |
|---|---|
| Length | 107 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.43 |
| Shannon entropy | 0.37224 |
| G+C content | 0.48850 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -28.33 |
| Energy contribution | -28.05 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
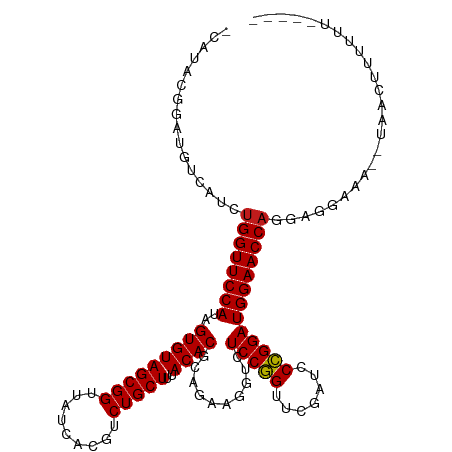
>dm3.chr3L 5349426 107 - 24543557 -----AAAAAAGUUA--CUUCCUCCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUGCCAAUCGCUUG- -----....((((..--....(..((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))..)......)))).- ( -32.40, z-score = -2.84, R) >anoGam1.chr2L 45432148 89 - 48795086 ------------------------CUGGUUCCAUCCAGGAUCGAACUGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCGCUCAUGCUCUCUCG-- ------------------------..((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))((....)).......-- ( -27.40, z-score = -2.22, R) >droGri2.scaffold_15110 7150282 105 - 24565398 ------CAAAAAGUA---AUCCACAUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAAUUGAGCGCCGAGUG- ------.........---...(((.(((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))).(((.....))))))- ( -29.70, z-score = -1.75, R) >droMoj3.scaffold_6680 11783997 105 - 24764193 ------AAAAAAAAA---ACCCACAUGGUUCCAUCCGGGAUCGAACCGGAGACCUUUUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUGCAAAACGUUUG- ------.......((---((.....(((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))))(((....))))))).- ( -27.80, z-score = -1.85, R) >droVir3.scaffold_13049 8121241 105 - 25233164 ------CAAAAGUAA---AGCCACUUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUGAGCACCGUCUG- ------.........---.((((.((((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))))).)).))........- ( -33.80, z-score = -2.63, R) >droWil1.scaffold_180698 7114872 115 + 11422946 CAAAAAAUUGACUCAACCAGAAUCCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUGAAAGCCGCAGCG ........................((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))......((....)). ( -31.70, z-score = -1.83, R) >droPer1.super_27 123157 107 - 1201981 ------AAAAAGUUA-AUUUCCUUCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGCUGAUGUCUGCAUU- ------.....((..-((.((...((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))..)).))..))...- ( -32.80, z-score = -2.65, R) >dp4.chrXR_group6 5000531 107 + 13314419 ------AAAAAGUUA-AUUUCCUUCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGCUGACGUCUGCAUU- ------.....((..-((.((...((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))..)).))..))...- ( -32.80, z-score = -2.55, R) >droAna3.scaffold_13337 10096500 109 + 23293914 -----AAAAAAGUUACUCAUCAACCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUGAACAUCGACUG- -----.....((((.....(((..((((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))))).))).....)))).- ( -33.80, z-score = -3.40, R) >droEre2.scaffold_4784 8040760 108 - 25762168 -----AAAAAAGCUA-CUUUCCUGCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUGCCAACCGAAUG- -----..........-........((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))..............- ( -31.40, z-score = -2.66, R) >droYak2.chr3L 5915726 108 - 24197627 -----AAAAAAGUUA-CUUUCCUGCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUGACAUUUGAAUG- -----..........-...((...((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))..))..........- ( -31.50, z-score = -2.75, R) >droSec1.super_2 5294685 107 - 7591821 -----AAAAAAGUUA--UUUCCUCCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUACCAUUCGUGUG- -----..........--.......((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))((((.....)))).- ( -32.00, z-score = -2.62, R) >droSim1.chr3L 4864024 107 - 22553184 -----AAAAAAGCUA--UUUCCUCCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUGCCAUUCGUUUG- -----....((((..--....(..((((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))..)......)))).- ( -31.60, z-score = -2.52, R) >consensus _____AAAAAAGUUA__UUUCCUCCUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUGACAUCCGUAUG_ .........................(((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))))................ (-28.33 = -28.05 + -0.28)

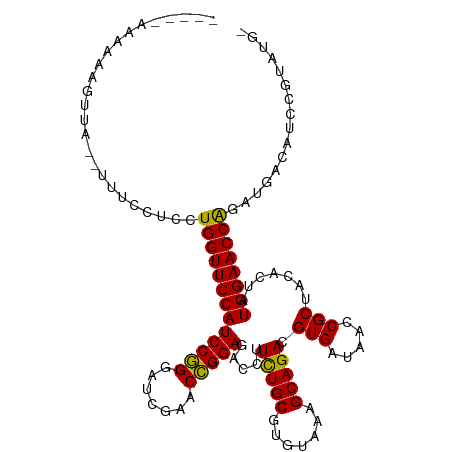
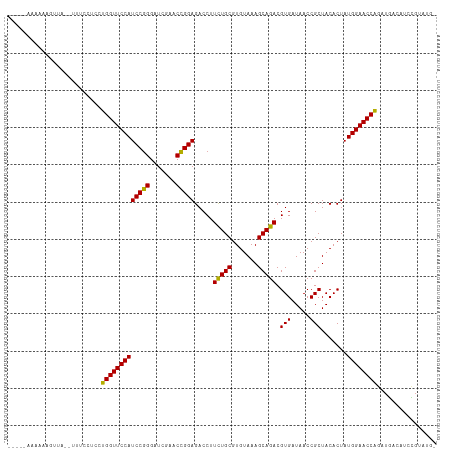
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:52 2011