| Sequence ID | dm3.chr3L |
|---|---|
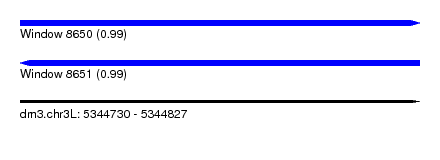
| Location | 5,344,730 – 5,344,827 |
| Length | 97 |
| Max. P | 0.992316 |

| Location | 5,344,730 – 5,344,827 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Shannon entropy | 0.42756 |
| G+C content | 0.33338 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -12.61 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992238 |
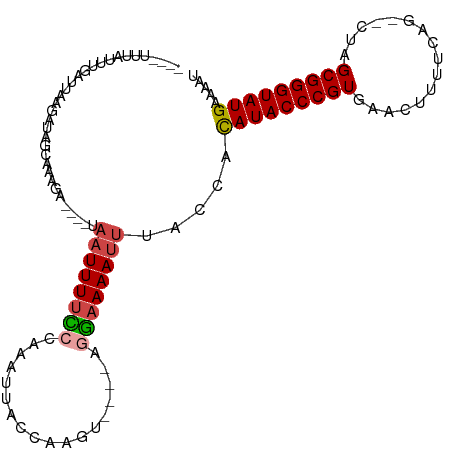
| Prediction | RNA |
Download alignment: ClustalW | MAF
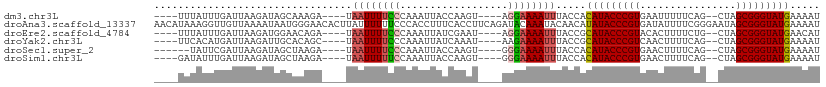
>dm3.chr3L 5344730 97 + 24543557 AUUUUCAUACCCGCUAG--CUGAAAAAUUCACGGGUAUGUGGUAAAUUUUCCU----ACUUGGUAAUUUGGGAAAAUUA----UCUUUGCUAUCUUAAUCAAAUAAA---- .....((((((((....--.(((.....)))))))))))((((((((((((((----(..(....)..)))))))))..----...))))))...............---- ( -20.90, z-score = -2.40, R) >droAna3.scaffold_13337 10091611 111 - 23293914 AUUUUCAUACCCGCUAUUCCCGAAAAUAUCACGGGUAUAUGUUGUAUUUGUAUCUGAAGGUGAAAGGUGGGAAAAAAUAAGUGUUCCCAUUAUUUUAACAACCUUUAUGUU .....((((((((((.(((((((.....)).((((((((.........))))))))..)).))).))))))..(((((((.((....))))))))).........)))).. ( -23.50, z-score = -0.84, R) >droEre2.scaffold_4784 8035787 97 + 25762168 AUGUUCAUACCCGCUAG--CAGAAAAGUGUACGGGUAUGCGGUAAAUUUUCCU----AUUCGAUAAUUUGGGAAAAUUA----UCUGUUCCAUCUUAAUCAAAUAAA---- .....((((((((...(--(......))...))))))))(((..(((((((((----(..........)))))))))).----.)))....................---- ( -23.50, z-score = -2.62, R) >droYak2.chr3L 5910629 97 + 24197627 AUUUUCAUACCCGCUAG--CUGAAAAGUUGACGGGUAUGCGGUAAAUUUUCUU----AUUUGAUAAUUUGGGAAAAUUA----GCUGUGCAAUCUUAAUCAUGUGAA---- ...((((((((((.(((--((....))))).)))))))(((((.(((((((((----(..(....)..)))))))))).----)))))................)))---- ( -29.70, z-score = -4.12, R) >droSec1.super_2 5289747 95 + 7591821 AUUUUCAUACCCGCUAG--CUGAAAAGUUCACGGGUAUGUGGUAAAUUUUCCC----ACUUGGUAAUUUGGGAAAAUUA----UCUUAGCUAUCUUAAUCGAAUA------ .....((((((((..((--((....))))..)))))))).(((((.(((((((----(..(....)..)))))))))))----))....................------ ( -29.10, z-score = -4.22, R) >droSim1.chr3L 4859032 97 + 22553184 AUUUUCAUACCCGCUAG--CUGAAAAGUUCACGGGUAUGUGGUAAAUUUUCCC----ACUUGGUAAUUUGGAAAAAUUA----UCUUAGCUAUCUUAAUCAAAUAUC---- ......(((((((..((--((....))))..)))))))((((.........))----))..((((((((....))))))----))......................---- ( -24.90, z-score = -3.66, R) >consensus AUUUUCAUACCCGCUAG__CUGAAAAGUUCACGGGUAUGUGGUAAAUUUUCCU____ACUUGAUAAUUUGGGAAAAUUA____UCUUUGCUAUCUUAAUCAAAUAAA____ .....((((((((..................)))))))).....(((((((((................)))))))))................................. (-12.61 = -13.06 + 0.45)

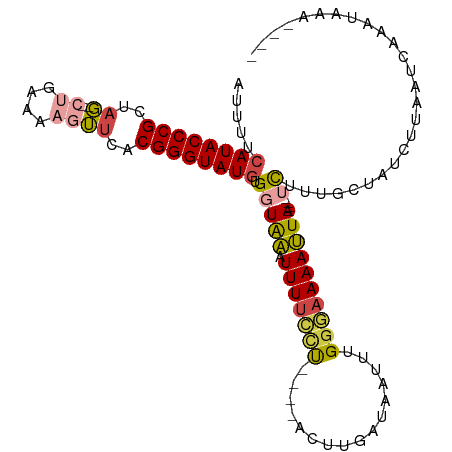
| Location | 5,344,730 – 5,344,827 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Shannon entropy | 0.42756 |
| G+C content | 0.33338 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5344730 97 - 24543557 ----UUUAUUUGAUUAAGAUAGCAAAGA----UAAUUUUCCCAAAUUACCAAGU----AGGAAAAUUUACCACAUACCCGUGAAUUUUUCAG--CUAGCGGGUAUGAAAAU ----.((((((.....))))))......----.((((((((.............----.)))))))).....(((((((((...........--...)))))))))..... ( -18.98, z-score = -2.47, R) >droAna3.scaffold_13337 10091611 111 + 23293914 AACAUAAAGGUUGUUAAAAUAAUGGGAACACUUAUUUUUUCCCACCUUUCACCUUCAGAUACAAAUACAACAUAUACCCGUGAUAUUUUCGGGAAUAGCGGGUAUGAAAAU .....((((((.(..(((((((((....)).)))))))..)..)))))).......................(((((((((..(((((....))))))))))))))..... ( -23.10, z-score = -1.44, R) >droEre2.scaffold_4784 8035787 97 - 25762168 ----UUUAUUUGAUUAAGAUGGAACAGA----UAAUUUUCCCAAAUUAUCGAAU----AGGAAAAUUUACCGCAUACCCGUACACUUUUCUG--CUAGCGGGUAUGAACAU ----((((((((((......((((....----.....))))......)))))))----)))...........(((((((((...(......)--...)))))))))..... ( -22.00, z-score = -2.71, R) >droYak2.chr3L 5910629 97 - 24197627 ----UUCACAUGAUUAAGAUUGCACAGC----UAAUUUUCCCAAAUUAUCAAAU----AAGAAAAUUUACCGCAUACCCGUCAACUUUUCAG--CUAGCGGGUAUGAAAAU ----...........((((((((...))----.))))))...(((((.((....----..)).)))))....((((((((..(.((....))--.)..))))))))..... ( -15.20, z-score = -1.34, R) >droSec1.super_2 5289747 95 - 7591821 ------UAUUCGAUUAAGAUAGCUAAGA----UAAUUUUCCCAAAUUACCAAGU----GGGAAAAUUUACCACAUACCCGUGAACUUUUCAG--CUAGCGGGUAUGAAAAU ------......................----.((((((((((..........)----))))))))).....(((((((((...((....))--...)))))))))..... ( -24.20, z-score = -3.48, R) >droSim1.chr3L 4859032 97 - 22553184 ----GAUAUUUGAUUAAGAUAGCUAAGA----UAAUUUUUCCAAAUUACCAAGU----GGGAAAAUUUACCACAUACCCGUGAACUUUUCAG--CUAGCGGGUAUGAAAAU ----..(((((.....))))).......----.((((((((((..........)----))))))))).....(((((((((...((....))--...)))))))))..... ( -22.00, z-score = -2.66, R) >consensus ____UUUAUUUGAUUAAGAUAGCAAAGA____UAAUUUUCCCAAAUUACCAAGU____AGGAAAAUUUACCACAUACCCGUGAACUUUUCAG__CUAGCGGGUAUGAAAAU .................................((((((((..................)))))))).....(((((((((................)))))))))..... (-14.51 = -14.84 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:51 2011