| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,342,824 – 5,342,966 |
| Length | 142 |
| Max. P | 0.735532 |

| Location | 5,342,824 – 5,342,966 |
|---|---|
| Length | 142 |
| Sequences | 7 |
| Columns | 157 |
| Reading direction | forward |
| Mean pairwise identity | 66.80 |
| Shannon entropy | 0.64842 |
| G+C content | 0.41792 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -11.60 |
| Energy contribution | -14.14 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
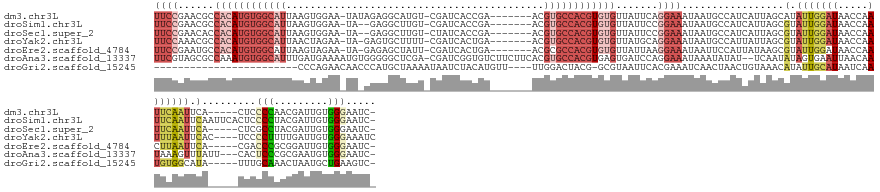
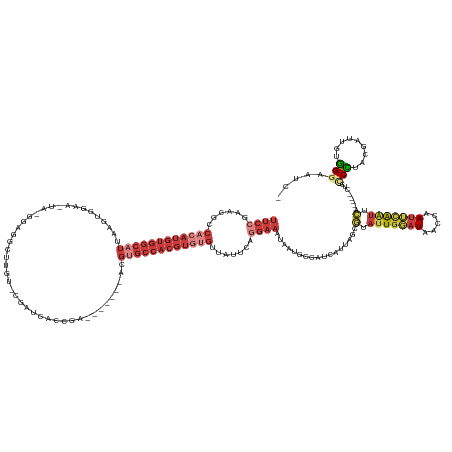
>dm3.chr3L 5342824 142 + 24543557 UUCCGAACGCCACAUGUGGCAUUAAGUGGAA-UAUAGAGGCAUGU-CGAUCACCGA-------ACGUGCCACGUGUGUUAUUCAGGAAAUAAUGCCAUCAUUAGCAUAUUGGAUAACCAAUUCAAUUCA-----CUCCCCAACGAUUGUGGGAAUC- ........((((....)))).....(((((.-......(((((((-((.....)).-------)))))))...((.(((((((((....(((((....))))).....)))))))))))......))))-----)((((((.....)).))))...- ( -40.80, z-score = -1.75, R) >droSim1.chr3L 4857091 145 + 22553184 UUCCGAACGCCACAUGUGGCAUUAAGUGGAA-UA--GAGGCUUGU-CGAUCACCGA-------ACGUGCCACGUGUGUUAUUCCGGAAAUAAUGCCAUCAUUAGCGUAUUGGAUAACCAAUUCAAUUCAAUUCACUCCCCUACGAUUGUGGGAAUC- .(((((((((...(((((((((((..(((((-((--(.(((.(((-((.....)).-------))).))).......))))))))....)))))))).)))..)))).))))).........................(((((....)))))....- ( -39.90, z-score = -1.17, R) >droSec1.super_2 5287852 140 + 7591821 UUCCGAACACCACAUGUGGCAUUAAGUGGAA-UA--GAGGCUUGU-CUAUCACCGA-------ACGUGCCACGUGUGUUAUUCCGGAAAUAAUGCCAUCAUUAGCGUAUUGGAUAACCAAUUCAAUUCA-----CUCGCCUACGAUUGUGGGAAUC- (((((.....((((((((((((....(((.(-((--((......)-))))..))).-------..))))))))))))......)))))....(.((((.(((.(((.(((((((.....)))))))...-----..)))....))).)))).)...- ( -40.90, z-score = -1.93, R) >droYak2.chr3L 5908690 143 + 24197627 UUCCAAACGCCACAUGUGGCAUUAACUAGAA-UA-GAGUGCUUUU-CGAUCACUGA-------ACGUGCCACGUGUGUUAUGCAGGAAAUAAUGCCAUUAUUAGCGUAUUGGAUAACCAAUUUAAUUCAC----UCCCCUUUUGAUUGUGGGAAAUC .(((((((((((((((((((((....(((..-..-(((.....))-).....))).-------..))))))))))))....(((........)))........)))).))))).................----(((((........).)))).... ( -37.90, z-score = -2.15, R) >droEre2.scaffold_4784 8033909 141 + 25762168 UUCCGAAUGCCACAUGUGGCAUUAAGUAGAA-UA-GAGAGCUAUU-CGAUCACUGA-------ACGCGCCACGUGUGUUAUUAAGGAAAUAAUUCCAUUAUAAGCGUAUUGGAUAACCAACUUAAUUCA-----CGACCCGCGGAUUGUGGGAAUC- .(((((((((((((((((((....(((.(((-((-(....)))))-)....)))..-------....)))))))))).......((((....)))).......)))).)))))................-----...(((((.....)))))....- ( -42.10, z-score = -3.18, R) >droAna3.scaffold_13337 10089883 150 - 23293914 UUCGUAGCGCCAAAUGUGGCAUUUGAUGAAAAUGUGGGGGCUCGA-CGAUCGGUGUCUUCUUCACGUGCCACGUGAGUGAUCCAGGAAAUAAAUAUAU--UCAAUAUAGUGAAUUAACAAUAAAGUUUAUU---CACUCCCGCGAAUGUGGGAAUC- (((....(((...(((((((((.(((.........(((((((((.-....))).)))))).))).)))))))))..))).....)))...........--........((((((.(((......))).)))---)))((((((....))))))...- ( -42.80, z-score = -1.71, R) >droGri2.scaffold_15245 10307705 122 + 18325388 ------------------------CCCAGAACAACCCAUGCUAAAAUAAUCUACAUGUU----UUGGACUACG-GCGUAAUUCACGAAAUCAACUAACUGUAAACAUAUUGCAUAAUCAAUGUGGCAUA-----UUUGCAAACUAAUGCUGAAGUC- ------------------------.((((((((......................))))----))))(((.((-((((..........................(((((((......)))))))(((..-----..)))......)))))).))).- ( -23.85, z-score = -1.57, R) >consensus UUCCGAACGCCACAUGUGGCAUUAAGUGGAA_UA_GGAGGCUUGU_CGAUCACCGA_______ACGUGCCACGUGUGUUAUUCAGGAAAUAAUGCCAUCAUUAGCGUAUUGGAUAACCAAUUCAAUUCA_____CUCCCCUACGAUUGUGGGAAUC_ ((((......((((((((((((...........................................)))))))))))).......)))).................(.(((((((.....))))))).).........(((.........)))..... (-11.60 = -14.14 + 2.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:48 2011