| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,335,175 – 5,335,274 |
| Length | 99 |
| Max. P | 0.918830 |

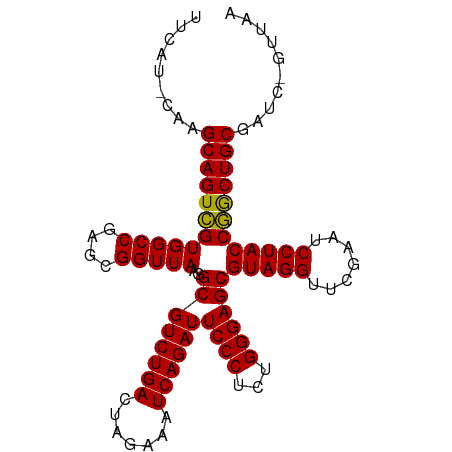
| Location | 5,335,175 – 5,335,274 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Shannon entropy | 0.25202 |
| G+C content | 0.53856 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -32.53 |
| Energy contribution | -32.14 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5335175 99 + 24543557 UUCAU-CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAUUAGUUAA .....-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).......... ( -32.90, z-score = -1.65, R) >droSim1.chr3L 4849471 98 + 22553184 UUCAU-CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAUC-GUUAA .....-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))....-..... ( -32.90, z-score = -1.45, R) >droSec1.super_2 5280218 98 + 7591821 UUCAU-CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAUC-GUUAA .....-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))....-..... ( -32.90, z-score = -1.45, R) >droYak2.chr3L 5900888 98 + 24197627 UUCAU-CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAUC-GUUAA .....-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))....-..... ( -32.90, z-score = -1.45, R) >droEre2.scaffold_4784 8026329 98 + 25762168 UUCGU-CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAUC-GUUGA ..((.-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))...)-).... ( -33.20, z-score = -0.84, R) >dp4.chr3 17013702 94 - 19779522 -UCCA-CAAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAUGAA---- -....-......((((..((((..........((((((((.......))))))((((...))))))(((((.......)))))))))..))))...---- ( -33.30, z-score = -1.26, R) >droPer1.super_2 6080617 94 - 9036312 -UCCA-CAAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAUGAA---- -....-......((((..((((..........((((((((.......))))))((((...))))))(((((.......)))))))))..))))...---- ( -33.30, z-score = -1.26, R) >droAna3.scaffold_13266 17922367 94 - 19884421 ------CAAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAAAGGUUCA ------..(((..(((..((((..........((((((((.......))))))((((...))))))(((((.......)))))))))..)))...))).. ( -33.60, z-score = -1.38, R) >droWil1.scaffold_181096 8516187 100 + 12416693 UUCACAGAAGCAGUUGUGGCCGAGCGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCAGCUGCGAGAAGUUGU .........((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).......... ( -31.10, z-score = -0.35, R) >droVir3.scaffold_12970 1725169 99 + 11907090 GUCAU-GGAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGACUGCGAAUGGGCUA (((..-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).....))).. ( -35.90, z-score = -1.12, R) >droMoj3.scaffold_6680 11843761 99 - 24764193 UUCAG-CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAAUGGAAAA ((((.-...((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))...))))... ( -34.40, z-score = -1.23, R) >droGri2.scaffold_15245 13573129 86 + 18325388 -------AAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAA------- -------......(((..((((..........((((((((.......))))))((((...))))))(((((.......)))))))))..))).------- ( -31.90, z-score = -1.49, R) >anoGam1.chr3L 6066541 99 - 41284009 UUCAC-GAAGCAGUCGUGGCCGAGUGGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGGUGUCGAAA (((((-(..((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))..))).))). ( -35.20, z-score = -1.03, R) >consensus UUCAU_CAAGCAGUCGUGGCCGAGCGGUUAAGGCGUCUGACUAGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGAUC_GUUAA .........((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......)))))))))))).......... (-32.53 = -32.14 + -0.39)

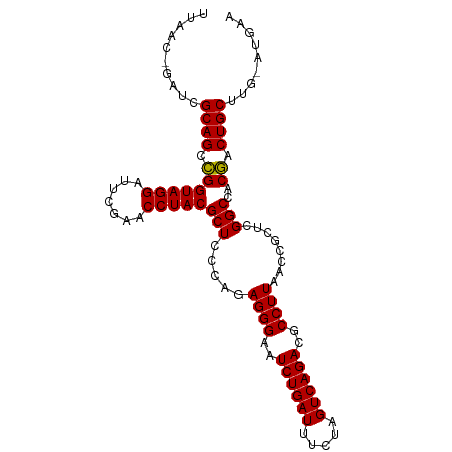
| Location | 5,335,175 – 5,335,274 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Shannon entropy | 0.25202 |
| G+C content | 0.53856 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -27.08 |
| Energy contribution | -26.94 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5335175 99 - 24543557 UUAACUAAUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG-AUGAA ..........((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))...-..... ( -32.02, z-score = -2.32, R) >droSim1.chr3L 4849471 98 - 22553184 UUAAC-GAUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG-AUGAA .....-....((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))...-..... ( -32.02, z-score = -1.99, R) >droSec1.super_2 5280218 98 - 7591821 UUAAC-GAUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG-AUGAA .....-....((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))...-..... ( -32.02, z-score = -1.99, R) >droYak2.chr3L 5900888 98 - 24197627 UUAAC-GAUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG-AUGAA .....-....((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))...-..... ( -32.02, z-score = -1.99, R) >droEre2.scaffold_4784 8026329 98 - 25762168 UCAAC-GAUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG-ACGAA ....(-(.((((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))..)-))).. ( -32.52, z-score = -1.87, R) >dp4.chr3 17013702 94 + 19779522 ----UUCAUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCUUG-UGGA- ----(((((.((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).))))..)-))))- ( -28.32, z-score = -0.59, R) >droPer1.super_2 6080617 94 + 9036312 ----UUCAUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCUUG-UGGA- ----(((((.((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).))))..)-))))- ( -28.32, z-score = -0.59, R) >droAna3.scaffold_13266 17922367 94 + 19884421 UGAACCUUUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCUUG------ ........(((..(((((((((.......)))))........((((..((((((.....))))))..)))).......))))..))).......------ ( -27.20, z-score = -1.13, R) >droWil1.scaffold_181096 8516187 100 - 12416693 ACAACUUCUCGCAGCUGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCGCUCGGCCACAACUGCUUCUGUGAA ........((((((..(((((((((((((.......((((...)))).......))))))))....(((..........))).....))))).)))))). ( -29.64, z-score = -1.15, R) >droVir3.scaffold_12970 1725169 99 - 11907090 UAGCCCAUUCGCAGUCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUCC-AUGAC .....(((..((((((((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)))))))...-))).. ( -32.12, z-score = -1.99, R) >droMoj3.scaffold_6680 11843761 99 + 24764193 UUUUCCAUUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG-CUGAA .....((.(((..(((((((((.......)))))((......((((..((((((.....))))))..))))....)).))))..))).))....-..... ( -29.30, z-score = -1.11, R) >droGri2.scaffold_15245 13573129 86 - 18325388 -------UUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCUU------- -------.(((..(((((((((.......)))))........((((..((((((.....))))))..)))).......))))..)))......------- ( -27.20, z-score = -1.50, R) >anoGam1.chr3L 6066541 99 + 41284009 UUUCGACACCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACCACUCGGCCACGACUGCUUC-GUGAA .............(((((((((.......)))))........((((..((((((.....))))))..)))).......))))(((((.....))-))).. ( -29.30, z-score = -0.96, R) >consensus UUAAC_GAUCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCUAGUCAGACGCCUUAACCGCUCGGCCACGACUGCUUG_AUGAA ..........((((.(((((((.......)))))(((.....((((..((((((.....))))))..))))........)))..)).))))......... (-27.08 = -26.94 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:45 2011