| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,313,339 – 5,313,436 |
| Length | 97 |
| Max. P | 0.854072 |

| Location | 5,313,339 – 5,313,436 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 64.61 |
| Shannon entropy | 0.63642 |
| G+C content | 0.41714 |
| Mean single sequence MFE | -19.97 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.49 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
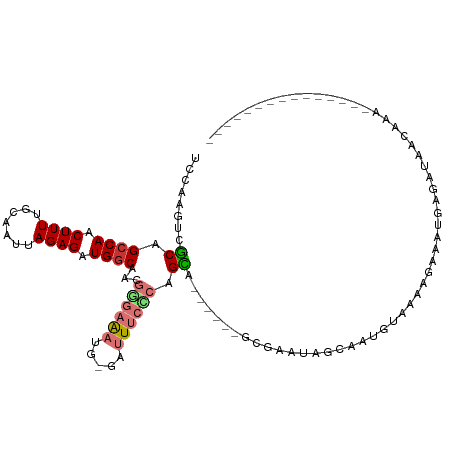
>dm3.chr3L 5313339 97 + 24543557 UCCAAGUCGCAGCCAACUUUUGCAAUUAGAGAUGGCAACGGGAAAUG-GUUUUCCCAGCAUCG---GCGAAUAGCAAUGUUAAAGAAAUGAGAUAACCCAA-------------- ((((..((((.((((.((((.......)))).))))...((((((..-..)))))).))....---((.....)).........))..)).))........-------------- ( -23.50, z-score = -1.21, R) >droPer1.super_27 81548 73 + 1201981 UCCAAGUCCCAGCCAACUUUCGCAAUUAGAGAUGGCAACAAUAGAGGCAAAUCUUCAGGG------AAAAGCGAUAGAA------------------------------------ ......((((.((((.((((.......)))).)))).......((((.....)))).)))------)............------------------------------------ ( -16.40, z-score = -1.63, R) >dp4.chrXR_group6 4959251 73 - 13314419 UCCAAGUCCCAGCCAACUUUCGCAAUUAGAGAUGGCAACAAUAGAGGCAAAUCUUCAGGG------AAAAGCGAUAGAA------------------------------------ ......((((.((((.((((.......)))).)))).......((((.....)))).)))------)............------------------------------------ ( -16.40, z-score = -1.63, R) >droAna3.scaffold_13337 10060134 93 - 23293914 UCCGAGUCUCAGCCAACUUUCGCAAUUAGAGAUGGCAACGGGAAAUG-AAUUUCUCAGGG------GCAUAUUUUAGCUUAGUAACUGAGAGUAAAUUAA--------------- .....(((((.((((.((((.......)))).))))...((((((..-..)))))).)))------)).((.((((.(((((...)))))..)))).)).--------------- ( -22.00, z-score = -1.12, R) >droEre2.scaffold_4784 8004006 109 + 25762168 UCCAAGUCGCCGCCAACUUUUGCAAUUAGAGAUGGCAACGGGAAAUG-GCUUUCACAGCAGCGCAUAGUAAAAGUAGUGCAAAAGAAAUGAGAUAACAAAUGUGCCAAAA----- (((....((..((((.((((.......)))).))))..)))))..((-((..........((((............))))...............((....))))))...----- ( -18.80, z-score = 1.48, R) >droYak2.chr3L 5872855 108 + 24197627 UCCAAGUCGCCGGCAACUUUUGCAAUUAGAGAUGGCAACGGGAAAUG-GCUUUCACAGCA------GCGAAUGGCAAUGUAAAAGAAAUGAGAUAACAAAUAUUAAAGAUAAAAA .(((..((.(((((..((((.......))))...))..)))))..))-)((((.(((...------((.....))..))).)))).............................. ( -15.60, z-score = 1.19, R) >droSec1.super_2 5258473 93 + 7591821 UCCAAGUCGCUGCCAACUUUUGCAAUUAGAGAUGGCAACGGGAAAUG-GUUUUCCCAGCA------GCGAAUAGCAAUGUAAAAGAAAUGAGAUAACAAA--------------- ((((..(((((((((.((((.......)))).))))...((((((..-..))))))))).------((.....)).........))..)).)).......--------------- ( -24.20, z-score = -2.15, R) >droSim1.chr3L 4827319 93 + 22553184 UCCAAGUCGCAGCCAACUUUUGCAAUUAGAGAUGGCAACGGGAAAUG-GCUUUCCCAGCA------GCGAAAAGCAAUGUAAAAGAAAUGAGAUAACAAA--------------- ((((..((((.((((.((((.......)))).))))...((((((..-..)))))).)).------((.....)).........))..)).)).......--------------- ( -22.90, z-score = -1.77, R) >consensus UCCAAGUCGCAGCCAACUUUUGCAAUUAGAGAUGGCAACGGGAAAUG_GAUUUCCCAGCA______GCGAAUAGCAAUGUAAAAGAAAUGAGAUAACAAA_______________ ........((.((((.((((.......)))).))))...((((((.....)))))).))........................................................ (-11.06 = -11.49 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:43 2011