| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,261,645 – 5,261,738 |
| Length | 93 |
| Max. P | 0.648630 |

| Location | 5,261,645 – 5,261,738 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Shannon entropy | 0.42753 |
| G+C content | 0.42141 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.68 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5261645 93 - 24543557 --AGGGCAGAAAGC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCCGCCG-------- --((((..((((((----((.(((((.(((.......)----)).)))))((..((((((((.....--.))))))))..))...)))--)))))..))))......-------- ( -30.50, z-score = -2.78, R) >droGri2.scaffold_15110 7061161 101 - 24565398 --AUACGGAAAUGUGGACGACGCCAUAUUGGCUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUACAGUGACCCCCAAAGAACACG------ --.(((.(((.((..((.((((((((.(((.......)----)).))))))))..(((((((.....--.)))))))))..)).)))...)))................------ ( -21.90, z-score = -0.38, R) >droMoj3.scaffold_6680 11675053 104 - 24764193 CUGGCAGGAAACGUGUACGACGCCAUAUUGGCUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUACUGUGUGACCCCAAGUAGAUCU----- .((((.(....)((.....))))))..((((...((((----(((((.(((...((((((((.....--.)))))))).))).))))..)))))....))))........----- ( -24.40, z-score = -0.08, R) >droVir3.scaffold_13049 8015381 103 - 25233164 AUUGCCGGAAACGUGUCCGACGCCAUAUUGGCUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUACUGUG-ACCCCCAAGCACGUCU----- ..(((..(((((.(((((((((((((.(((.......)----)).))))))))......)))))..)--)))).....(..((......))..-).......))).....----- ( -24.50, z-score = -0.06, R) >droWil1.scaffold_180698 6985053 105 + 11422946 --AGUACAGG--------GACGCCAUAUUGGCUAACAUAUACAAAAUGGCGUUUAUGAUGGGCAAAGUUUUUGAUCAUUUGCAAUUUGAAAUGUUGACCCAAGCCCGUUCUCACA --......((--------((((((((.(((...........))).)))))))......(((((((.(((((.(((........))).))))).))).))))..)))......... ( -26.80, z-score = -1.07, R) >dp4.chrXR_group6 8411047 93 - 13314419 -----AGGGGAAGC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG-UUUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCUUCCAGC------ -----(((((((((----((.(((((.(((.......)----)).)))))((..((((((((.....-..))))))))..))...)))--))))...))))........------ ( -26.20, z-score = -1.71, R) >droAna3.scaffold_13337 10016904 97 + 23293914 --AGGGCAGAAAGCUACGGACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCCACAU-------- --((((..((((((...(((((((((.(((.......)----)).)))))))))((((((((.....--.)))))))).........)--)))))..))))......-------- ( -30.10, z-score = -2.96, R) >droEre2.scaffold_4784 7958347 93 - 25762168 --AGGACAGAAAGC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCCUCCG-------- --(((...((((((----((.(((((.(((.......)----)).)))))((..((((((((.....--.))))))))..))...)))--)))))...)))......-------- ( -26.40, z-score = -2.39, R) >droYak2.chr3L 5826335 93 - 24197627 --AGGACAGAAAGC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCCUCUG-------- --(((...((((((----((.(((((.(((.......)----)).)))))((..((((((((.....--.))))))))..))...)))--)))))...)))......-------- ( -26.40, z-score = -2.43, R) >droSec1.super_2 5214332 93 - 7591821 --AGGGCAGAAAGCU----ACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCCUCCG-------- --((((..((((((.----..(((((.(((.......)----)).)))))((..((((((((.....--.))))))))..)).....)--)))))..))))......-------- ( -28.80, z-score = -2.76, R) >droSim1.chr3L 4779477 93 - 22553184 --AGGGCAGAAAGC----GACGCCAUAUUGGAUAACAU----AAAAUGGCGUUUAUGAUGGGCAAAG--UUUCAUCAUUUGCAAUUUG--CUUUUGACCCUCCUCCG-------- --((((..((((((----((.(((((.(((.......)----)).)))))((..((((((((.....--.))))))))..))...)))--)))))..))))......-------- ( -30.50, z-score = -3.10, R) >consensus __AGGACAGAAAGC____GACGCCAUAUUGGAUAACAU____AAAAUGGCGUUUAUGAUGGGCAAAG__UUUCAUCAUUUGCAAUUUG__CUUUUGACCCUCCUCCG________ .....................(((((...................)))))((..((((((((........))))))))..))................................. (-13.59 = -13.68 + 0.09)

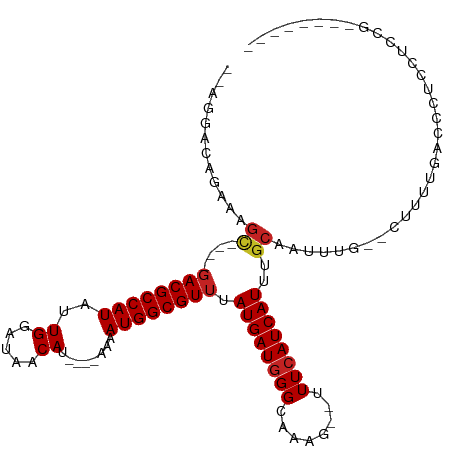
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:42 2011