| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,260,039 – 5,260,132 |
| Length | 93 |
| Max. P | 0.660463 |

| Location | 5,260,039 – 5,260,132 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 70.96 |
| Shannon entropy | 0.54661 |
| G+C content | 0.51113 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.660463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
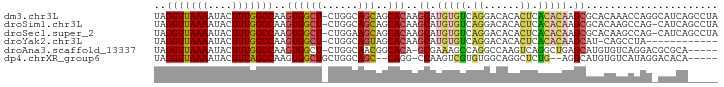
>dm3.chr3L 5260039 93 + 24543557 UAGGUUAAAAUACUUUGGCCAAGUGGCU-CUGGCAGCAGCACAAGCAUGUGUCAGGACACACUCACACAAGCGCACAAACCAGGCAUCAGCCUA ..(((((((....))))))).(((((..-((((((.((((....)).))))))))..).))))..................((((....)))). ( -26.80, z-score = -1.12, R) >droSim1.chr3L 4777893 92 + 22553184 UAGGUUAAAAUACUUUGGCCAAGUGGCU-CUGGCAGCAGCACAAGCAUGUGUCAGGACACACUCACACAAGCGCACAAGCCAG-CAUCAGCCUA ..(((((((....)))))))....((((-(((((....((....((.(((((.((......)).))))).))))....)))))-....)))).. ( -28.50, z-score = -1.45, R) >droSec1.super_2 5212777 92 + 7591821 UAGGUUAAAAUACUUUGGCCAAGUGGCU-CUGGAAGCAGCACAAGCAUGUGUCAGGACACACUCACACAAGCGCACAAGCCAG-CAUCAGCCUA ..(((((((....)))))))....((((-((((.....((....((.(((((.((......)).))))).)))).....))))-....)))).. ( -26.00, z-score = -0.91, R) >droYak2.chr3L 5824736 80 + 24197627 UAGGUUAAAAUACUUUGGCCAAGUGGCU-CUGGCAGUAGCACAAGCAUGUGUCAGGACACACUCACACAAGCAU-CAGCCUA------------ ..(((((((....)))))))....((((-...((....((....)).(((((.((......)).))))).))..-.))))..------------ ( -22.80, z-score = -1.16, R) >droAna3.scaffold_13337 10015353 87 - 23293914 UAGGUUAAAAUACUUUGGCCAAGUGGCU-CUGGCAACGGCACA-GCGAAAGCCAGGCCAAGUCAGGCUGAGCAUGUGUCAGGACGCGCA----- ..(((((((....)))))))..((((..-((((((...((...-((....))..((((......))))..))...))))))..).))).----- ( -32.60, z-score = -1.62, R) >dp4.chrXR_group6 4902567 84 - 13314419 UAGGUUAAAAUACUUUAGCCAAGUGGCUGCUGGCAGC--CAGG-CCAAGUCGUGUGGCAGGCUCUG--AGGCAUGUGUCAUAGGACACA----- ..(((((((....)))))))...((((((....))))--))..-....(((.(((((((.(((...--.)))...))))))).)))...----- ( -32.50, z-score = -1.78, R) >consensus UAGGUUAAAAUACUUUGGCCAAGUGGCU_CUGGCAGCAGCACAAGCAUGUGUCAGGACACACUCACACAAGCACACAACCCAG_CAUCA_____ ..(((((((....)))))))..((.(((......))).))....((.(((((.((......)).))))).))...................... (-13.16 = -13.55 + 0.39)
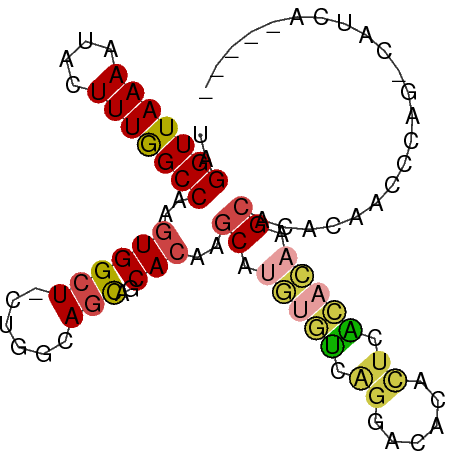

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:40 2011