
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,250,491 – 5,250,594 |
| Length | 103 |
| Max. P | 0.625957 |

| Location | 5,250,491 – 5,250,594 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Shannon entropy | 0.26725 |
| G+C content | 0.40779 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -19.08 |
| Energy contribution | -18.16 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5250491 103 + 24543557 AUUGGCUCGGAACUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGUAAAACCC--AAGGAUAAUGCAUGAGUUGGUCAGGUUAC-------CCUGAC--AAAUCUGGCUAAG .(((((.((((.....((((((((((..((((............))))..)))).--...(......)..))))))((((((....-------))))))--...))))))))). ( -29.00, z-score = -2.54, R) >droSim1.chr3L 4769501 103 + 22553184 AUUGGCUCGGAACUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGUAAAACCC--AAGGAUAAUGCAUGAGUUGGUCAGGUUAC-------CCUGAC--AAAUCUGGCUAAU ((((((.((((.....((((((((((..((((............))))..)))).--...(......)..))))))((((((....-------))))))--...)))))))))) ( -29.00, z-score = -2.63, R) >droSec1.super_2 5203220 103 + 7591821 AUUGGCUCGGAACUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGUAAAACCC--AAGGAUAAUGCAUGAGUUGGUCAGGUUAC-------CCUGAC--AAAUCUGGCUAAU ((((((.((((.....((((((((((..((((............))))..)))).--...(......)..))))))((((((....-------))))))--...)))))))))) ( -29.00, z-score = -2.63, R) >droYak2.chr3L 5815286 103 + 24197627 AUUGGCUUGGAACUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGUAAAACCC--AAGGAUAAUGCAUGAGUUGGUCAGGUUAC-------CCUGAC--AAAUCUGGCUAAU (((((((.(((.....((((((((((..((((............))))..)))).--...(......)..))))))((((((....-------))))))--...)))))))))) ( -26.10, z-score = -1.69, R) >droEre2.scaffold_4784 7945801 103 + 25762168 AUUGGCUCGGAACUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGUAAAACCC--AAGGAUAAUGCAUGAGUUGGUCAGGUUAC-------CCUGAC--AAAUCUGGCUAAU ((((((.((((.....((((((((((..((((............))))..)))).--...(......)..))))))((((((....-------))))))--...)))))))))) ( -29.00, z-score = -2.63, R) >droAna3.scaffold_13337 10006491 112 - 23293914 AUUGGCUCGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAGAACCC--AAGGAUAAUGCAUGAGUUGGCCAGGUUAGGACUGAACCUGACUGAAAUCUGGCUAAU ((((((.((((..((.((((((((((..((((............))))..)))).--...(......)..))))))..))(((((((......)))))))....)))))))))) ( -30.90, z-score = -2.24, R) >dp4.chrXR_group6 4889680 108 - 13314419 AUUGGCUCGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAAAAACCCAAAGGAUAAUGCAUGACUUGGUCAGGUUUUGU----GUCUGAC--AAAUCUGGCUAAU .((((((...((((((....)))))).)))))).............(((....((....))....))).....((((((((((((.((----.....))--)))))))))))). ( -23.80, z-score = -0.83, R) >droPer1.super_27 14329 108 + 1201981 AUUGGCUAGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAAAAACCCAAAGGAUAAUGCAUGACUUGGUCAGGUUUUGU----GUCUGAC--AAAUCUGGCUAAU .((((((...((((((....)))))).)))))).............(((....((....))....))).....((((((((((((.((----.....))--)))))))))))). ( -23.80, z-score = -0.84, R) >droVir3.scaffold_13049 7995519 107 + 25233164 AUUGGCUCGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAGAGACC---AGGAUAAUGCAUG-GCUGGCCAGACCCUGGUUACCUCUGC---AAAUCUGGCUAAU ((((((.((((.(((((......)))))..................(((((((((---(((.....(..((-(....)))..)))))))...))))))---...)))))))))) ( -32.30, z-score = -2.03, R) >droMoj3.scaffold_6680 11659299 107 + 24764193 AUUGGCUCGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAGAGACC---AGGAUAAUGCAUG-GCUGGCCAGACGCUGGUUACCUCUGC---AAAUCUGGCUAAU ((((((.((((.(((((......)))))..................(((((((((---((.....((..((-(....)))..)))))))...))))))---...)))))))))) ( -29.90, z-score = -1.36, R) >droGri2.scaffold_15110 7047572 105 + 24565398 AUUGGCUCGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAAA--CC---AGGAUAAUGCAUG-GCUGGCCAGACGUUGGUUACCUCUGC---AAAUCUGGCUAAU ((((((.((((........(..((((..((((............)))).))--))---..)....((((.(-(.(((((((...))))))).)).)))---)..)))))))))) ( -27.00, z-score = -1.03, R) >consensus AUUGGCUCGGAAUUGACAACUCGGUUAAGCUAACAAUAUUUGAAUAGCAAAACCC__AAGGAUAAUGCAUGAGUUGGUCAGGUUACG______CCUGAC__AAAUCUGGCUAAU .((((((...((((((....)))))).)))))).............(((....((....))....)))......((((((((......................)))))))).. (-19.08 = -18.16 + -0.92)

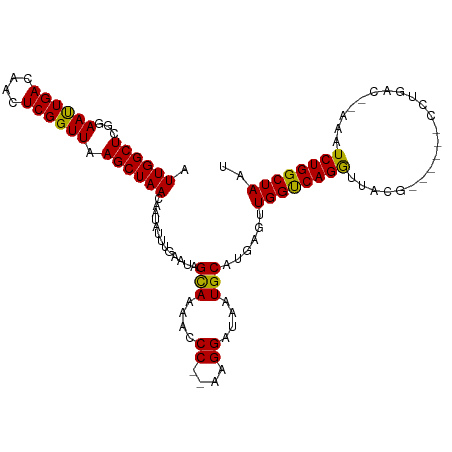
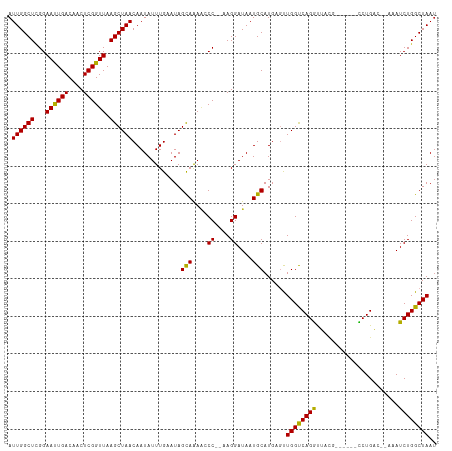
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:39 2011