
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 5,242,909 – 5,243,027 |
| Length | 118 |
| Max. P | 0.860200 |

| Location | 5,242,909 – 5,243,027 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.76 |
| Shannon entropy | 0.07363 |
| G+C content | 0.38030 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.87 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 5242909 118 + 24543557 AAAUUCGCAUAAAAAUAAUAAAGCUUUUGCCACAUAACUCCCUAAUUUAUGCGAAUGCCCCUCUCUCAGAAGGUGUCAACUUUAUGAUAUAAUCCCAGCCAACAUGAAGCUAAUCCCC ..(((((((((((.........((....))....((......)).)))))))))))((((.((.....)).)).))...(((((((................)))))))......... ( -16.79, z-score = -1.44, R) >droSim1.chr3L 4761943 118 + 22553184 AAAUUCGCAUAAAAAUAAUAAAGCUUUUGCCACAUAACUCCCUAAUUUAUGCGAAUGCCCCUCUCUCAGAAGGUGUCAACUUUAUGAUAUAAUCCCAGCCAACAUGAAGCUAAUCCCC ..(((((((((((.........((....))....((......)).)))))))))))((((.((.....)).)).))...(((((((................)))))))......... ( -16.79, z-score = -1.44, R) >droSec1.super_2 5195781 117 + 7591821 AAAUUCGCAUAAAAAUAAUAAAGCGUUUGCCACAUAACUCCCUAAUUUAUGCGA-UGCCCCUCUCUCAGAAGGUGUCAACUUUAUGAUAUAAUCCUAGCCAACAUAAAGCUAAUCCCC ......(((((((.........((....))....((......)).)))))))((-((((..((.....)).))))))..(((((((................)))))))......... ( -15.29, z-score = -1.04, R) >droYak2.chr3L 5807497 118 + 24197627 AAAUUCGCAUAAAAAUAAUAAAGCUUUUGCCACAUAACUCCCUAAUUUAUGCGAAUGCCCCUCCCUCUAAAGGUGUCAACUUUAUGAUAUAAUCCCAGCGAACAUGAAGCUAAUCCCC ..(((((((((((.........((....))....((......)).)))))))))))................((((((......))))))......(((.........)))....... ( -16.40, z-score = -1.62, R) >droEre2.scaffold_4784 7938164 118 + 25762168 AAAUUCGCAUAAAAAUAAUAAAGCUUUUGCCACAUAACUGCCUAAUUUAUGCGAAUGCCCCUCCCUCAAAAGGUGCCAACUUUAUGAUAUAAUCCCAACGAACAUGAAGCUAAUCCCC ..(((((((((((.........((....))..((....)).....)))))))))))((.(((........))).))...(((((((................)))))))......... ( -18.69, z-score = -2.48, R) >consensus AAAUUCGCAUAAAAAUAAUAAAGCUUUUGCCACAUAACUCCCUAAUUUAUGCGAAUGCCCCUCUCUCAGAAGGUGUCAACUUUAUGAUAUAAUCCCAGCCAACAUGAAGCUAAUCCCC ..(((((((((((.........((....))....((......)).)))))))))))((.(((........))).))...(((((((................)))))))......... (-15.99 = -15.87 + -0.12)


| Location | 5,242,909 – 5,243,027 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.76 |
| Shannon entropy | 0.07363 |
| G+C content | 0.38030 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
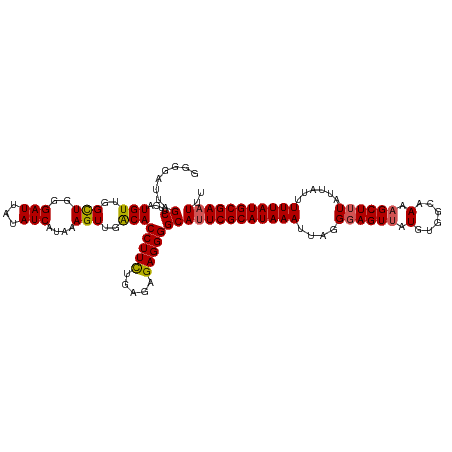
>dm3.chr3L 5242909 118 - 24543557 GGGGAUUAGCUUCAUGUUGGCUGGGAUUAUAUCAUAAAGUUGACACCUUCUGAGAGAGGGGCAUUCGCAUAAAUUAGGGAGUUAUGUGGCAAAAGCUUUAUUAUUUUUAUGCGAAUUU ........((....(((..(((..(((...)))....)))..)))(((((.....)))))))(((((((((((....((((((.(......).))))))......))))))))))).. ( -30.70, z-score = -1.93, R) >droSim1.chr3L 4761943 118 - 22553184 GGGGAUUAGCUUCAUGUUGGCUGGGAUUAUAUCAUAAAGUUGACACCUUCUGAGAGAGGGGCAUUCGCAUAAAUUAGGGAGUUAUGUGGCAAAAGCUUUAUUAUUUUUAUGCGAAUUU ........((....(((..(((..(((...)))....)))..)))(((((.....)))))))(((((((((((....((((((.(......).))))))......))))))))))).. ( -30.70, z-score = -1.93, R) >droSec1.super_2 5195781 117 - 7591821 GGGGAUUAGCUUUAUGUUGGCUAGGAUUAUAUCAUAAAGUUGACACCUUCUGAGAGAGGGGCA-UCGCAUAAAUUAGGGAGUUAUGUGGCAAACGCUUUAUUAUUUUUAUGCGAAUUU ........((....(((..(((..(((...)))....)))..)))(((((.....))))))).-(((((((((.(((((.(((........))).))))).....))))))))).... ( -28.70, z-score = -1.70, R) >droYak2.chr3L 5807497 118 - 24197627 GGGGAUUAGCUUCAUGUUCGCUGGGAUUAUAUCAUAAAGUUGACACCUUUAGAGGGAGGGGCAUUCGCAUAAAUUAGGGAGUUAUGUGGCAAAAGCUUUAUUAUUUUUAUGCGAAUUU ........(((((.((((.(((..(((...)))....))).))))(((....)))..)))))(((((((((((....((((((.(......).))))))......))))))))))).. ( -28.00, z-score = -1.50, R) >droEre2.scaffold_4784 7938164 118 - 25762168 GGGGAUUAGCUUCAUGUUCGUUGGGAUUAUAUCAUAAAGUUGGCACCUUUUGAGGGAGGGGCAUUCGCAUAAAUUAGGCAGUUAUGUGGCAAAAGCUUUAUUAUUUUUAUGCGAAUUU ........(((((.(.((((..(((....(..((......))..))))..)))).).)))))(((((((((((....(((....)))(((....)))........))))))))))).. ( -26.00, z-score = -0.60, R) >consensus GGGGAUUAGCUUCAUGUUGGCUGGGAUUAUAUCAUAAAGUUGACACCUUCUGAGAGAGGGGCAUUCGCAUAAAUUAGGGAGUUAUGUGGCAAAAGCUUUAUUAUUUUUAUGCGAAUUU ........((....(((..(((..(((...)))....)))..)))(((((.....)))))))(((((((((((....((((((.(......).))))))......))))))))))).. (-26.12 = -26.16 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:04:37 2011